Pre-Requisites
A working setup of openCGA is required to setup a Testing environment. If user hasn't yet set it up, please follow the steps on installation guide and set it up.
Download Test Data
In order to populate the environment with real life data, User can download data from the following Genomes ftp. For this tutorial, we will download and use ALL.chr22.phase3_shapeit2_mvncall_integrated_v5a.20130502.genotypes.vcf.gz
User can use any other data, the setup process will remain same.
Initialisation Script
Download the initialisation.sh script. All of the following steps assume, user is under openCGA installation directory (/opt/opencga/). Below you can find explanation of initialisation script step by step:
This CLI command will create the database, the collections and all the indexes, it also creates the admin user with the specified password. The MongoDB database host and name are read from the /conf/catalog-configuration.yml file by default.
./bin/opencga-admin.sh catalog install -p <<< admin_P@ssword
Then user need to start catalog dameon
./opencga-admin.sh catalog daemon --start -p <<< admin_P@sswor
This following command will create a user name "John Doe" and ID "test". Note that as by default OpenCGA is configured as private which means that only admin user can create other users. We are using opencga-admin CLI
./bin/opencga-admin.sh users create -p -u test --user-email test@gel.ac.uk --user-name "John Doe" --user-password user_P@ssword <<< admin_P@sswor
Now we will use this newly created user "test" for further actions, for this first user need to login. The next statement will do that:
./bin/opencga.sh users login -u test -p <<< user_P@ssword
This will create a hidden directory in your home called .opencga. This directory will contain a file named ~/.opencga/session.json with the users and the session id, this will be used automatically by opencga.sh, this is valid only for some minutes, by doing this users do not have to write the password too many times. The contents of session.json file will look like :
{
"userId" : "test",
"sessionId" : "bvQGV1XOA1xt38Ae4meh",
"login" : "2016-12-15T14:02:58.981",
"logout" : null,
"timestamp" : 1481816675284
}
Now with new user, we create a project name "Reference studies GRCh37" and alias "reference_grch37" with the following command :
./opencga.sh projects create -a reference_grch37 -n "Reference studies GRCh37"
Next step, create a study name "" inside project "reference_grch37"
./opencga.sh studies create -a 1kG_phase3 -n "1000 Genomes Project - Phase 3" --project-id reference_grch37
Now lets link/register downloaded file(s)s with newly created study. This process will add a file entry in catalog with some information and stats of file(s)
./opencga.sh files link -i ALL.chr22.phase3_shapeit2_mvncall_integrated_v5a.20130502.genotypes.vcf.gz -s 1kG_phase3
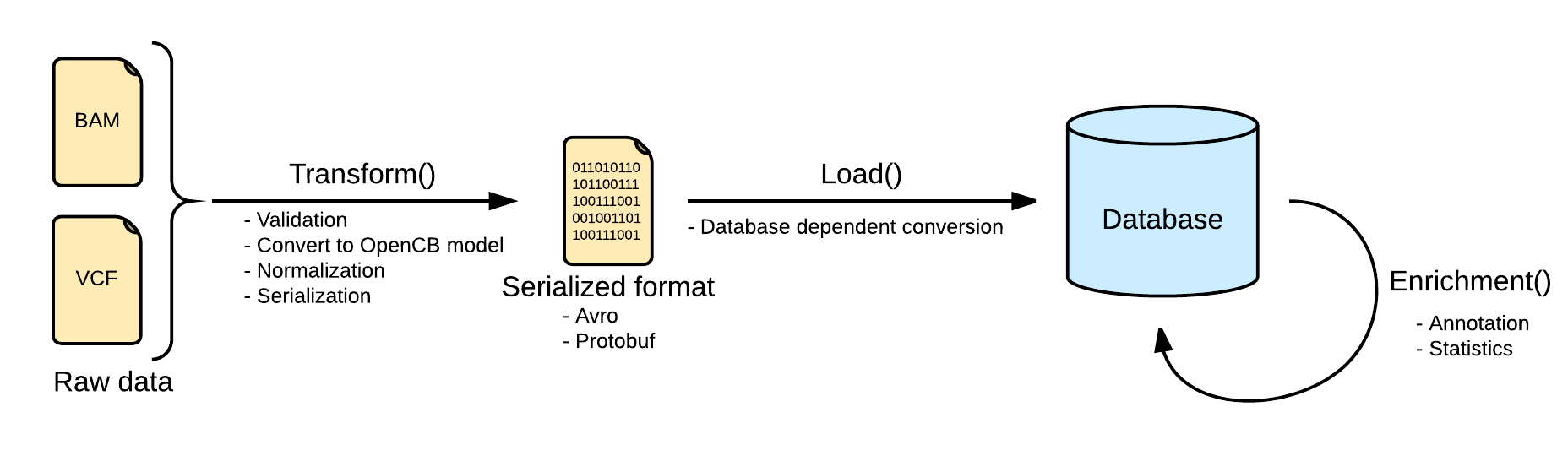
Now as this file is linked into openCGA catalog, user can index variants. Below is the pictorial representation of indexing pipeline:
The next step would be to transform this variant file. (Please have a look on wiki for detailed explanation of these steps)
./opencga.sh files index --id ALL.chr22.phase3_shapeit2_mvncall_integrated_v5a.20130502.genotypes.vcf.gz --transform --of JSON
Next we load transformed data into openCGA storage
./opencga.sh files index --id ALL.chr22.phase3_shapeit2_mvncall_integrated_v5a.20130502.genotypes.vcf.gz --load --of JSON
After this user can annotate the variants. We will use opencga-analysis.sh script for this purpose
mkdir -p /tmp/temporal_annotation ### temporary directory ./opencga-analysis.sh variant annotate -s 1kG_phase3 -o /tmp/temporal_annotat
As last step, use can calculate statistics on this data using the following command
mkdir -p /tmp/temporal_statistics ./opencga-analysis.sh variant stats -s 1kG_phase3 -o /tmp/temporal_statistics --cohort-ids ALL
For user ease, openCGA provides a single command to perform full pipeline operation. The following command can be executed in place of above four ones to achieve same results :
./opencga.sh variant index --file-id ALL.chr22.phase3_shapeit2_mvncall_integrated_v5a.20130502.genotypes.vcf.gz --calculate-stats --annotate
At this point, data is fully loaded into openCGA storage along with annotations and calculated stats. User can perform different query to access/analyse this data.
./bin/opencga-analysis.sh variant query --return-study 1kG_phase3 --region 1:14558108-14558112