OpenCGA benchmark is a rich test suite to benchmark different storage engine currently supported with OpenCGA for variant storage.
Execution Mode
Benchmark supports the following execution mode :
- Fixed
- Random
Fixed Mode
Its a fixed set of queries written in a YML file, benchmark will take each query (default) or a selection of queries passed as IDs arguments in --query, -q option and execute these as a certain number of users (-c, - -concurrency) for a specific number of time (-r, --num-repetition). Common parameters to each query are placed in baseQuery. A sample of fixedQuery is displayed below:
---
baseQuery :
summary : true
queries :
- id : "RegionAndBiotype"
description : "Purpose of this query"
query :
region : "22:16052853-16054112"
gene : "BRCA2"
biotype : "coding"
populationFrequencyMaf : "1kG_phase3:ALL>0.1"
tolerationThreshold : 300
- id : "Region"
description : "Purpose of this query"
query :
region : "22:16052853-16054112"
tolerationThreshold : 400
.....
sessionIds :
- ""
- ""
Following command will execute ALL queries written in fixedQueries.yml file as 10 users, five times each on REST server specified in "storage-configuration.yml" :
opencga-storage-admin.sh benchmark variant --concurrency 10 --num-repetition 5 --mode FIXED --connector REST
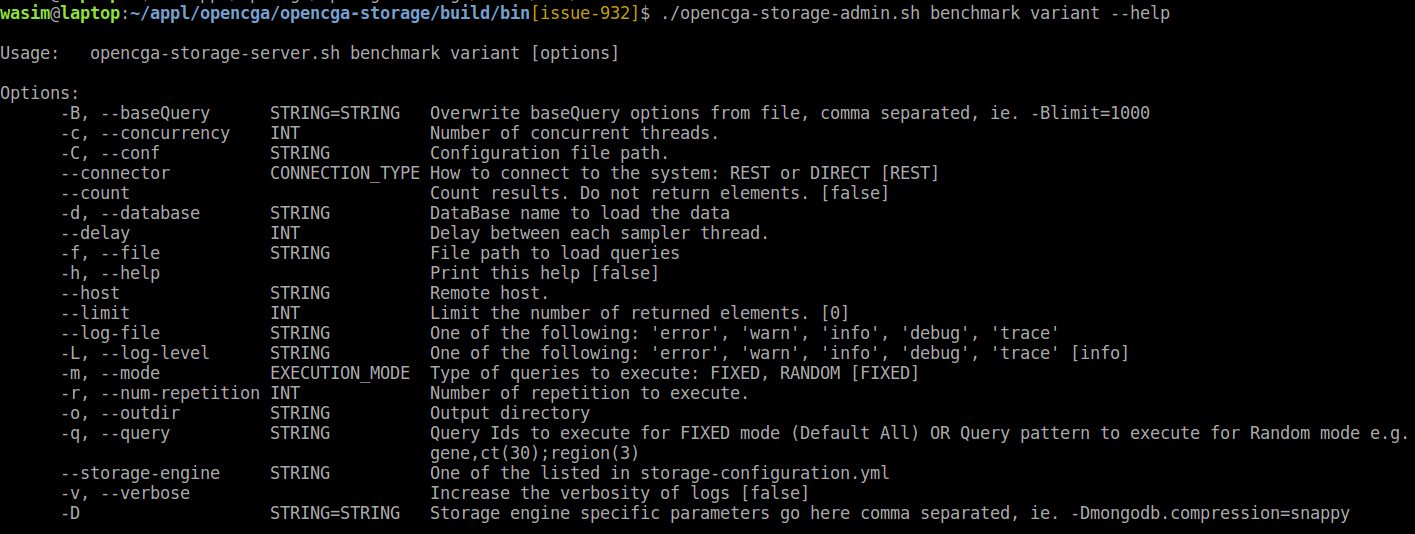
Complete list of options, default values and explanations can be displayed using - - help option from benchmark script :
Random Mode
Random mode supports creation of random queries from meta data provided in "randomQueries.yml" and execute these on selected storage engine :
---
baseQuery :
summary : true
exclude : studies
regions :
- chromosome : "1"
start : 1
end : 249250621
gene :
- DKFZP434A062
- GPSM1
ct : []
type :
- "SV"
- "CNV"
study :
- "1kG_phase3"
...
functionalScore :
- id : "cadd_raw"
min : 0
max : 1
- id : "cadd_scaled"
min : -10
max : 40
populationFrequencies :
- id : "1kG_phase3:ALL"
min : 0
max : 0.2
- id : "1kG_phase3:AFR"
min : 0
max : 0.15
proteinSubstitution :
- id : "polyphen"
min : 0.1
max : 0.9
operators : [">", "<"]
- id : "sift"
min : 0.1
max : 0.9
qual :
id : "polyphen"
min : 1
max : 9
operators : [">"]
conservation :
id : "phylop"
min : 0
max : 1
operators : ["=", "!="]
sessionIds :
- ""
- ""
Following command will generate two queries one with two different "ct" values and a gene value and second with a region value provided in "randomQueries.yml" file and execute as 10 users, five times each on REST server:
opencga-storage-admin.sh benchmark variant --concurrency 10 --num-repetition 5 --mode RANDOM -q "ct(2),gene;region"
Connection Type
Storage Engine
The following Storage engines are currently supported with OpenGCA :
- Mongo
- HBase