Versions Compared
Key
- This line was added.
- This line was removed.
- Formatting was changed.
Client-Server Model
User Interfaces
Higher level interfaces to HGVA include the web applicationHGVA is implemented following a Client-Server architecture. The server uses OpenCB OpenCGA project to load and index variant datasets (VCF and gVCF files), OpenCGA server provides a complete REST API to query metadata and variants. The client side implements three different user interfaces. First, a rich web-based data mining application based on OpenCB IVA project. Second, three client libraries for Java, Python and Javascript. Third, a command-line interface. Client libraries and command-line can query both metadata and variants and are part of OpenCGA project.
 Image Added
Image Added
OpenCGA Server
OpenCGA is an open-source project that aims to provide a Big Data storage engine and analysis framework for genomic scale data analysis of hundreds of terabytes. It implements different components:
- Catalog to store metadata
- Variant storage engine to provide real-time queries to big data in genomics. This can use MongoDB or HBase together with Solr. A Redis server is also used to cache queries.
- A complete RESTful API and gRPC for variant and alignment (BAM) streaming
- Client libraries and command-line to query data
HGVA uses a small cluster for OpenCGA installation. This consist of three servers for MongoDB and a single Solr server. HAProxy is used to balance queries to two Tomcat servers.
Client User Interfaces
Client user interfaces to HGVA include a rich web application based on IVA, client libraries in Java, Python and JavaScript, and a Command Line Interface. All of these make intensive use of HGVA's RESTful APIweb services (taken from OpenCGA), which is are accessible through an HAProxy load balancer. Currently, two production Tomcat machines run the RESTful API. gRPC servers are currently under development and will soon be enabled to improve performance. The RESTful API makes use of a Java API to query either Catalog or the Variants Storage. Catalog module stores and enables managing projects metadata, i.e.: data files, samples data, populations, etc. Variants Storage module allows managing and querying genomic variants data. Currently MongoDB is used as the data base management system; a replica set with three MongoDB servers supports HGVA installation. A Solr server is currently being use under develop branch. Likewise, a Redis server provides a cache that speeds-up expensive and frequent queries. Image Removed.
Image Removed.
OpenCB IVA
IVA is a highly customisable web application for Interactive Variant Analysis (IVA). It consists of several tools, HGVA activates two of them: Variant Browser and Facets. You can execute complex queries in IVA using any variant annotation including full-text search for disease descriptions.
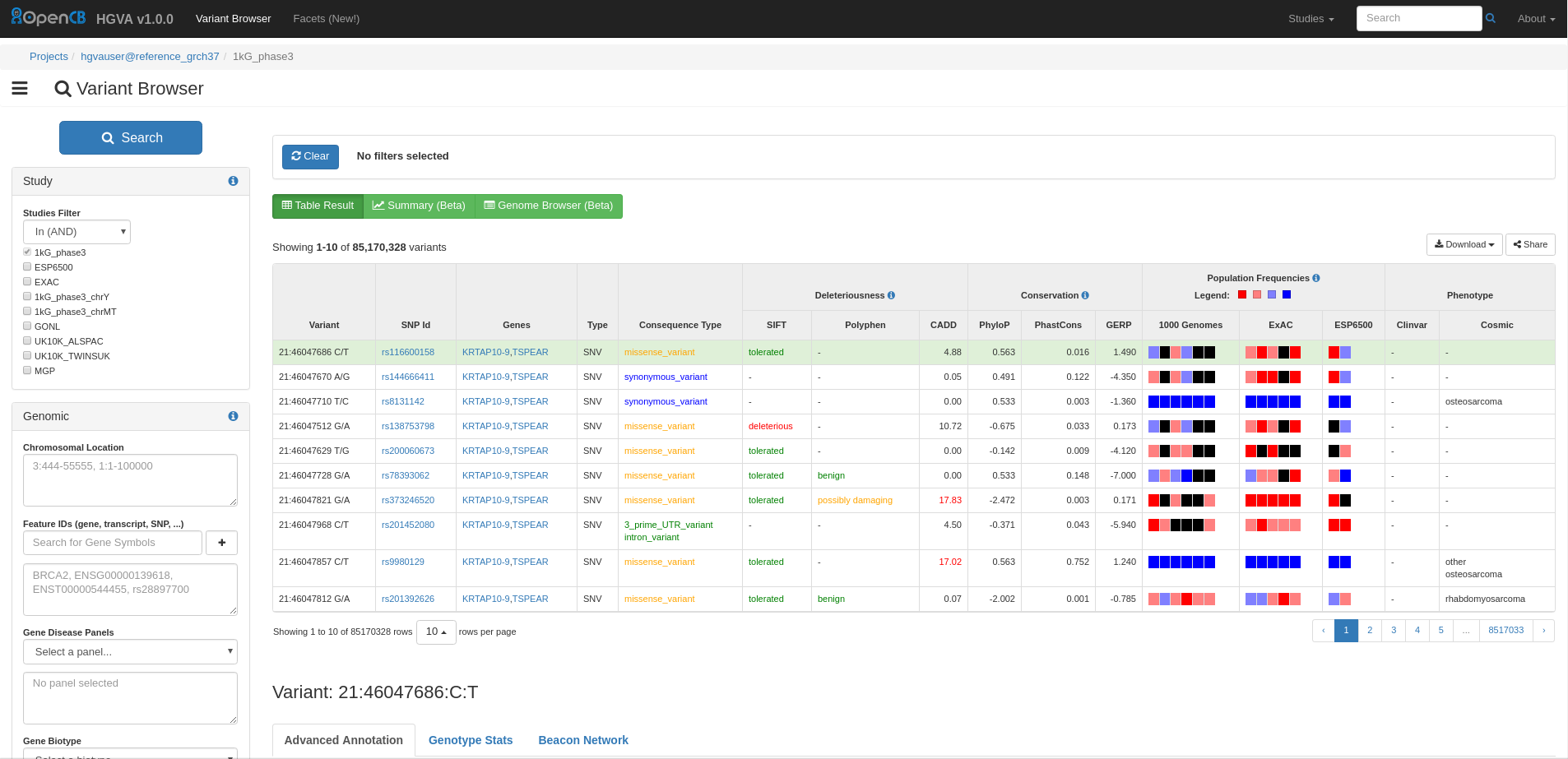 Image Added
Image Added
With Facets you can perform different aggregations of data:
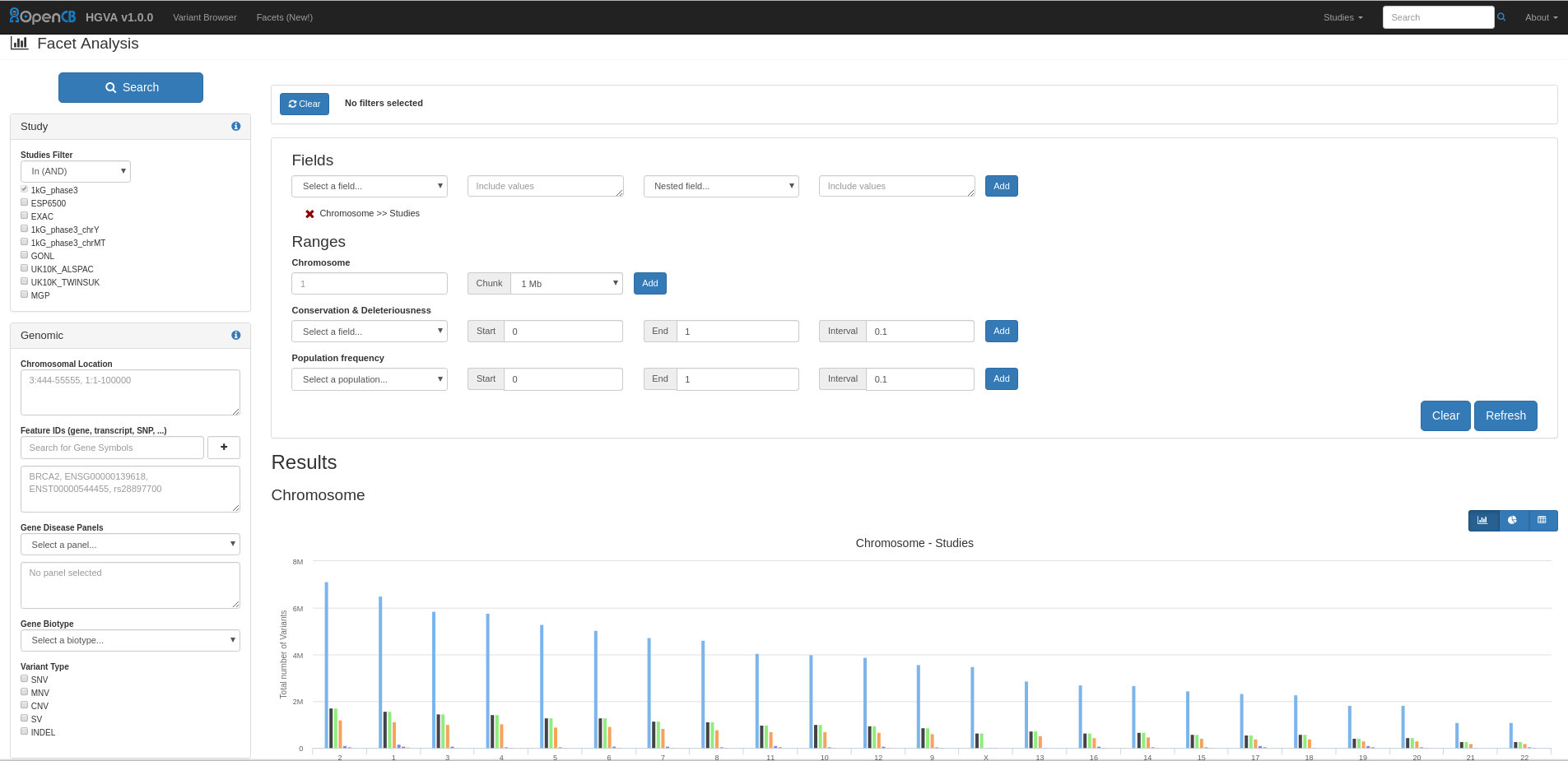 Image Added
Image Added
Table of Contents:
| Table of Contents | ||
|---|---|---|
|