Genomic and Clinical Data
Genomic variants of 4,700 genomes were loaded and indexed in OpenCGA. In total we loaded almost 5,000 VCF files accounting for about 20TB of compressed disk space.
For this proof of concept (PoC) we used the development version OpenCGA v2.0.0-beta using the Hadoop Variant Storage Engine that uses Apache HBase as back-end. We also used CellBase 4.6 for the variant annotation.
For the platform we used a 10-nodes Azure HDInsight 3.6 cluster using Data Lake Storage Gen2. HDInsight 3.6 uses Hortonworks HDP 2.6.5 (with Hadoop 2.7.3 and HBase 1.1.2) and we used Azure Batch for loading concurrently all the VCF files which had been copied previously to a NFS server, you can see details here:
| Node Type | Nodes | Azure Type | Cores | Memory (GB) | Storage |
|---|
| Hadoop Master | 3 | Standard_D12_V2 | 4 | 28 | Data Lake Gen2 |
| Hadoop Worker | 10 | Standard_DS13_V2 | 8 | 56 | Data Lake Gen2 |
| Azure Batch Queue | 10 | Standard_D4s_v3 | 4 | 16 | NFS Server |
We evaluated the new HDInsight 4 but after finding few issues and we decided to use the more stable HDInsight 3.6 (HDI3.6) over Data Lake Gen2 (DL2), we will refer to this as HDI3.6+DL2. We worked with Azure engineers to debug and fix these issues during the PoC, unfortunately we did no have time to repeat the benchmark.
As you will below in the analysis benchmark, once we completed the PoC we repeated some tests with 20 working nodes to study the performance improvement.
Table size
| Table | Compression | Size (TB) |
|---|
| Variants table | GZ | 2.9 |
| Variants table | SNAPPY | 4.7 |
Genomics Data Loading and Indexing
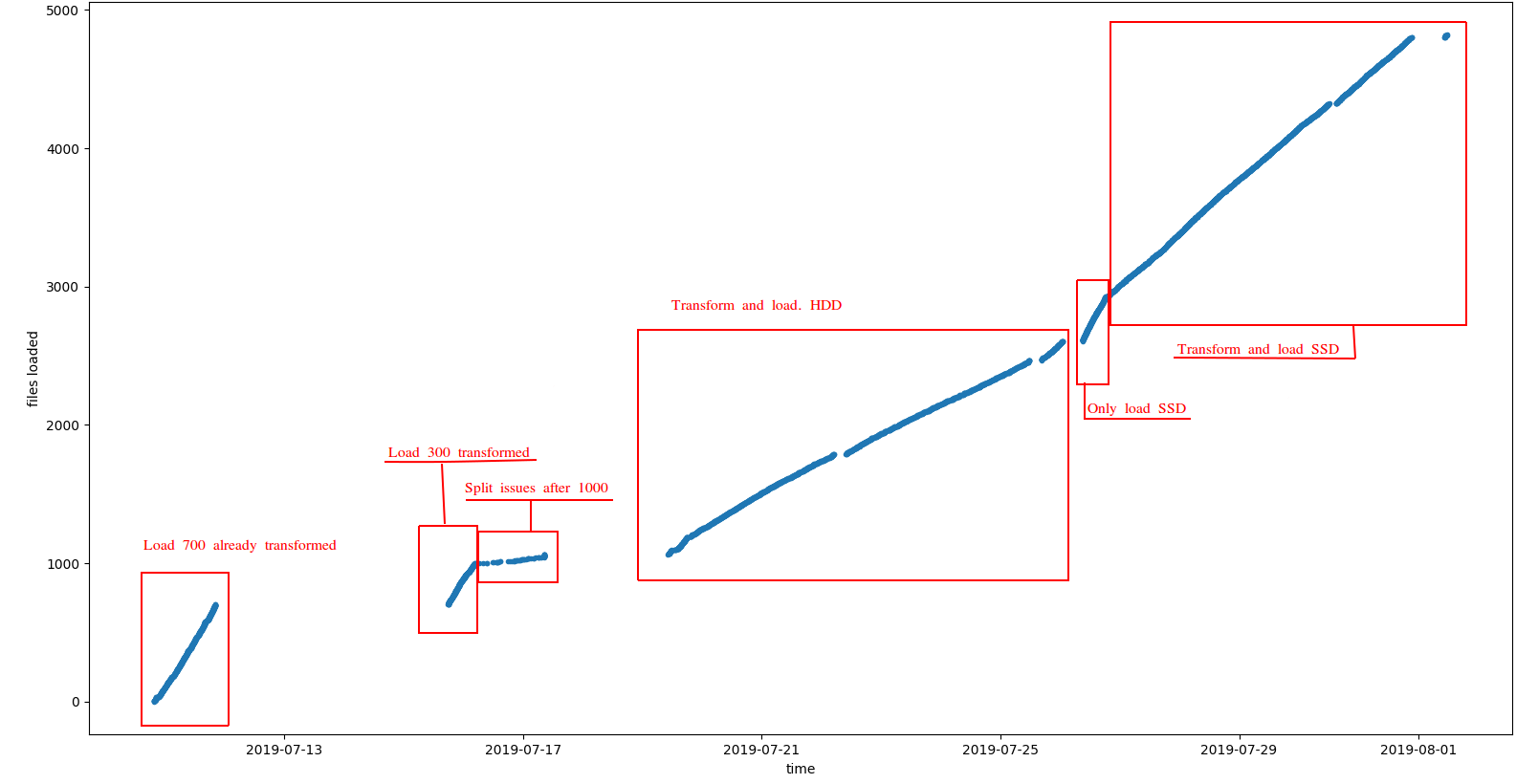
Number of loaded files across time. We can differentiate some sections with different performance.
The more representative section is the last one, where we upgraded the input disk to speed up the reading. In average, with the improved disk, processing up to 20 files simultaneously we have these numbers:
| Time | Time/nodes |
|---|
| Transform | 00:29:36 | 00:01:28 |
|---|
| Load | 00:46:19 | 00:02:19 |
|---|
| Total | 1:15:55 | 00:03:48 |
|---|
Index speed:
- 15.8 files/h
- 379.4 files/day
- 79.0 GB/h
- 1.85 TB/day
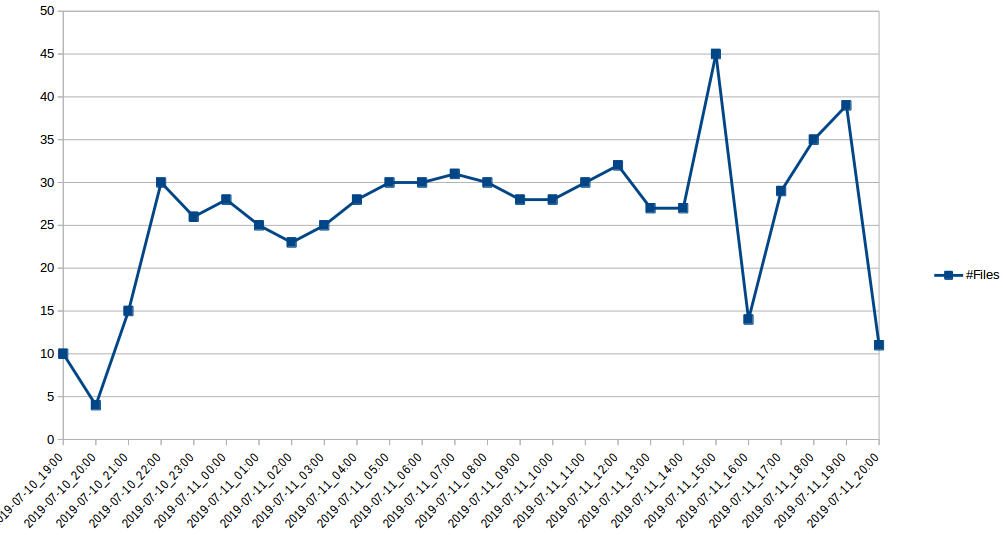
#Files Day Hour
10 2019-07-10 19
4 2019-07-10 20
15 2019-07-10 21
30 2019-07-10 22
26 2019-07-10 23
28 2019-07-11 00
25 2019-07-11 01
23 2019-07-11 02
25 2019-07-11 03
28 2019-07-11 04
30 2019-07-11 05
30 2019-07-11 06
31 2019-07-11 07
30 2019-07-11 08
28 2019-07-11 09
28 2019-07-11 10
30 2019-07-11 11
32 2019-07-11 12
27 2019-07-11 13
27 2019-07-11 14
45 2019-07-11 15
14 2019-07-11 16
29 2019-07-11 17
35 2019-07-11 18
39 2019-07-11 19
11 2019-07-11 20
Operations
First batch of 700 files
74.096.015 variants
Aggregate
Prepare: 529.303s [ 00:08:49 ]
Aggregate: 9591.626s [ 02:39:52 ]
Write: 7012.733s [ 01:56:53 ] -> Size : 59.5 GiB
Stats
1352.675s [ 00:22:33 ]
Annotate
Prepare: 722.327s [ 00:12:02 ]
Annot: 50384.383s [ 13:59:44 ]
Load: 28204.666s [ 07:44:04 ]
SampleIndex: 12403.542s [ 03:26:44 ]
Secondary index (Solr)
.....
Analysis Benchmark
Query and Aggregation Stats
Stats
GWAS
Clinical Analysis