Versions Compared
Key
- This line was added.
- This line was removed.
- Formatting was changed.
Understanding the URL
| Code Block | ||||
|---|---|---|---|---|
| ||||
http://host_url/webservices/rest/{apiVersion}/{resource}/{id(s)}/{endpoint}?{options} |
Where host_url is the name of the host machine and the name of war file deployed in web server (tomcat) over that machine for example bioinfodev.hpc.cam.ac.uk/opencga
Entities inside the { curly braces } are web service parameters, treated as variables.
| Code Block | ||||
|---|---|---|---|---|
| ||||
http://bioinfodev.hpc.cam.ac.uk/opencga/webservices/rest/v1/users/opencga/login?password=opencga |
As is explained later in this documentation, this RESTful web service will login user opencga.
- apiVersion (v1) : indicates OpenCGA version to retrieve information from, data models and API may change between versions.
- resource (users) : specifies the data type of what the user wants to retrieve in the id field. This can be one of resources listed below.
- id (opencga) : the resource id we want to query.
- endpoint (login) : these parameters must be specified depending on the nature of your input data. For example, if we want to query all files by a specific study (e.g. 1000genomes) we should use the studies resource and files endpoint.
- options (password) : variables in key value pair form, passed as query parameters.
APIVersion
ApiVersions are numbered as v1, v2, etc. At this moment we are heading to first stable apiVersion which is v1. However, when more apiVersions are available in the future the latest stable apiVersion will be always coded as latest.
Resources and Endpoints
There are 16 different resources implemented:
| Category | Description | Main Endpoints |
|---|---|---|
| users | Different methods to work with users | info, create, login, ... |
| projects | projects are defined for each user and contains studies | info, create, studies, ... |
| studies | studies are the main component of catalog, the can be shared and contain files, samples and jobs | info, create, files, samples, jobs, variants, alignments, groups, ... |
| files | files are added to the study and can be indexed to be queried | info, create, index, share, ... |
| jobs | jobs are tool executions that can be queued | info, create, ... |
| families | families are connected collection of individuals based on relationship | info, create, ... |
| individuals | samples come from the individuals | info, create, ... |
| samples | samples are each of the experiment samples, typically matches a NGS BAM file or VCF sample | info, create, annotate, share, ... |
| cohorts | these model a group of samples that share some common properties, these are used for data analysis | info, create, stats, samples, ... |
| clinical Analysis | this handles creating and search of a clinical analysis | info, create, ... |
| variableSet | variables annotate samples with different information useful for data analysis | info, crate, ... |
| analysis alignmentweb services to facilitate the alignment process | Operations over Read Alignments to facilitate complete analysis with different tools. | index, query, stats, coverage |
| analysis variant | web services Operations over Genomic Variants to facilitate complete variantanalysis usingwith different handy methodstools. | index, stats, query, validate, ibs, facet, samples, metadata |
| analysis tool | bioinformatics tools installed for data analysis | execute |
| Meta | gives the meta information about OpenCGA installation instance | ping, about, status |
| GA4GH | GA4GH standard web services to search genomics data in OpenCGA | variant search, reads search, responses |
IDs
This is the query parameter and the type matches resources path parameter, this is a unique identifier for the resource we are interacting. Plural means a list of IDs can be passed to improve performance with a single REST call separated by commas rather than multiple calls. OpenCGA preserves the order of the results with corresponding IDs. A Boolean variable, silent, can be set to indicate either if user is interested to receive partial results (true) as well or only expects complete set of results or nothing in case of any failure (resource doesn't exist, permission denied etc). As a trade off between performance and ease of use a maximum of 100 IDs are allowed in one web service.
Options
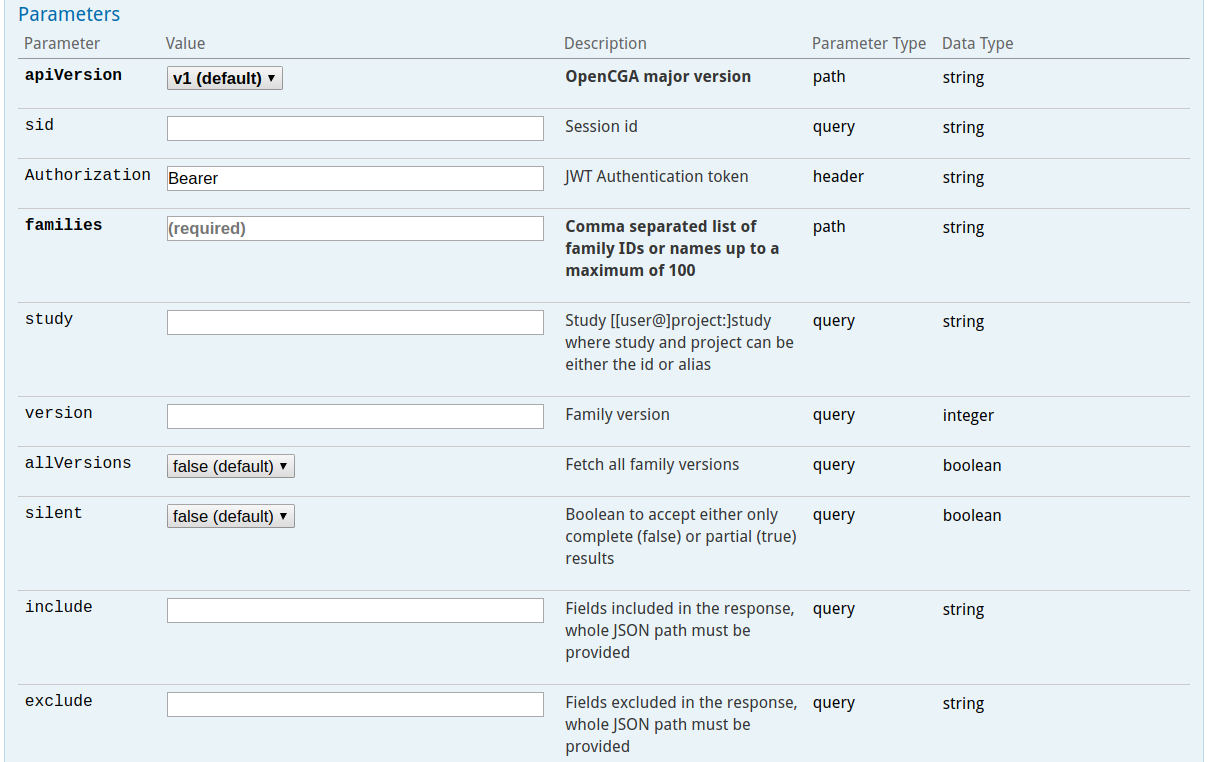
These query parameters can modify the behaviour of the query (exclude, include, limit, skip and count) or add some filters to some specific endpoints to add useful functionality. The following image shows some typical options for a certain web service.
Swagger
OpenCGA has been documented using Swagger project. Detailed information about resources, endpoints and options is available at:
http://bioinfodev.hpc.cam.ac.uk/opencga/webservices/
Client Libraries
Currently OpnnCGA supports the following client liberalise:
Deprecation Policy
Certain APIs are deprecated over the period of time as OpenCGA is a live project and continuously improved and new features are implemented. Deprecation cycle consists of a warning period to let make user aware that these services are considered for change and highly likely will be replaced followed by a deprecated message. OpenCGA supports deprecated services for two releases (Deprecated and Next one). Deprecated services are hidden in the following release of deprecation and finally removed completely in next release.
| Code Block | ||||
|---|---|---|---|---|
| ||||
Warning (working) ---> Deprecated (working) ---> Hidden (working) ---> Removed (not working) |
All deprecated web services are clearly marked as deprecated along with a warning help message for user.
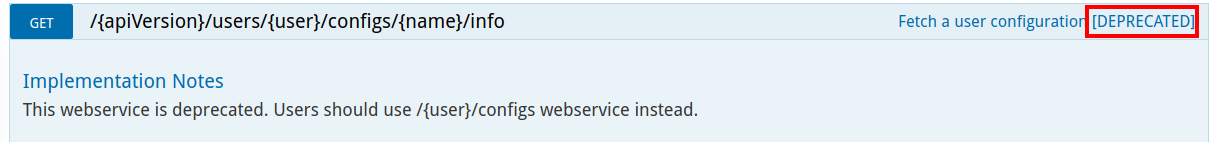
Table of Contents:
| Table of Contents | ||
|---|---|---|
|