Versions Compared
Key
- This line was added.
- This line was removed.
- Formatting was changed.
Index
Annotate
Custom annotation
Calculate Statistics
Define cohorts
Remove
Import
Export
Export statistics
Export statistics is an special case of export. Instead of export full variants, only the variant cohort statistics are exported.
As for variants export, there are multiple possible output formats:
VCF : Standard VCF format without samples information, with the stats as values in the INFO column.
| Code Block | ||||
|---|---|---|---|---|
| ||||
##fileformat=VCFv4.2
##FILTER=<ID=.,Description="No FILTER info">
##FILTER=<ID=PASS,Description="Valid variant">
##INFO=<ID=AC,Number=A,Type=Integer,Description="Total number of alternate alleles in called genotypes, for each ALT allele, in the same order as listed">
##INFO=<ID=AF,Number=A,Type=Float,Description="Allele Frequency, for each ALT allele, calculated from AC and AN, in the range (0,1), in the same order as listed">
##INFO=<ID=AN,Number=1,Type=Integer,Description="Total number of alleles in called genotypes">
#CHROM POS ID REF ALT QUAL FILTER INFO
22 16050115 . G A . PASS AC=1;AF=0.001;AN=1000
22 16050213 . C T . PASS AC=1;AF=0.001;AN=1000
22 16050319 . C T . PASS AC=1;AF=0.001;AN=1000
22 16050607 . G A . PASS AC=2;AF=0.002;AN=1000
|
TSV (Tab Separated Values). Simple format with each cohort in one column.
| Code Block | ||||
|---|---|---|---|---|
| ||||
#CHR POS REF ALT ALL_AN ALL_AC ALL_AF ALL_HET ALL_HOM
22 16050213 C T 1000 1 0.001 0.002 0.0
22 16050607 G A 1000 2 0.002 0.004 0.0
22 16050740 A - 1000 1 0.001 0.002 0.0
22 16050840 C G 1000 13 0.013 0.026 0.0
22 16051075 G A 1000 2 0.002 0.004 0.0
22 16051249 T C 1000 91 0.091 0.162 0.01
22 16051453 A C 998 74 0.074 0.144 0.004
22 16051453 A G 926 2 0.002 0.144 0.004
22 16051723 A - 1000 12 0.012 0.024 0.0
22 16051816 T G 1000 2 0.002 0.004 0.0
|
Operations
Variant Storage Operations are responsible for leaving variant data ready for querying and analysis, for instance VCF loading, integrity checks, sample genotype aggregation, indexing, or variant annotation are examples of operations. Operations can only be executed by admin users. Many operations write and update indexed data, this will significantly improve the quality and performance of different queries and analysis.
The OpenCGA Variant Storage Engine supports several operations to work with variant datasets:
Index Pipeline
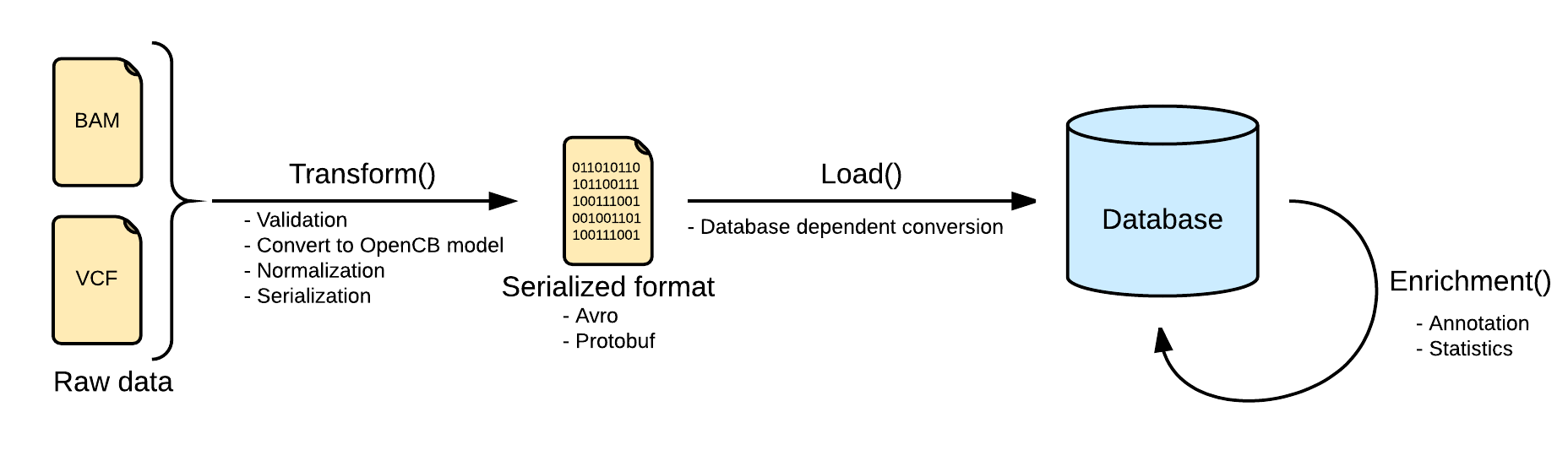 Image Added
Image Added
Table of Contents:
| Table of Contents | ||
|---|---|---|
|