Versions Compared
Key
- This line was added.
- This line was removed.
- Formatting was changed.
Index
Indexing variants does not apply any modification to the generic pipeline. The input file format is VCF, accepting different variations like gVCF or aggregated VCFs
Transform
Files are converted Biodata models. The metadata and the data are serialized into two separated files. The metadata is stored into a file named <inputFileName>.file.json.gz serializing in json a single instance of the biodata model VariantSource, which mainly contains the header and some general stats. Along with this file, the real variants data is stored in a file named <inputFileName>.variants.avro.gz with a set of variant records described as the biodata model Variant.
VCF files are read using the library HTSJDK, which provides a syntactic validation of the data. Further actions on the validation will be taken, like duplicate or overlapping variants detection.
By default, malformed variants will be skipped and written into a third optional file named <inputFileName>.malformed.txt . If the transform step generates this file, a curation process should be taken to repair the file. Otherwise, the variants would be skipped.
All the variants in the transform step will be normalized as defined here: Variant Normalization. This will help to unify the variants representation, since the VCF specification allows multiple ways of referring to a variant and some ambiguities.
Load
Loading variants from multiple files into a single database will effectively merge them. In most of the scenarios, with a good normalization, merging variants is strait forward. But in some other scenarios, with multiple alternates or overlapping variants, the merge requires more logic. More information at Merge Mode.
Details about load are dependent on the implementation.
Annotate
As part of the enrichment step, some extra information can be added to the variants database as Annotations. This VariantAnnotation can be fetch from Cellbase or read from local file provided by the user. The model of the variant annotation is defined in the project Biodata, in variantAnnotation.avdl
Custom annotation
The VariantAnnotation model includes a field for adding extra annotation attributes. This field is intended to contain custom annotation provided by the end user.
Additional attributes can be grouped by source. Each source will contain a set of key-value attributes creating this structure:
| Code Block | ||||
|---|---|---|---|---|
| ||||
VariantAnnotation = {
// ...
"additionalAttributes" : {
"<source1>" : {
"attribute" : {
"<key1>":"<value>",
"<key2>":"<value>",
"<key3>":"<value>"
}
},
"<source2>" : {
"attribute" : {
"<key1>":"<value>",
"<key2>":"<value>",
"<key3>":"<value>"
}
}
}
|
OpenCGA Storage is able to load this custom annotation from 3 different formats: GFF, BED and VCF. When loading the new annotation data, the user has to provide a name for the new custom annotation. Because the structure of these file formats is slightly different, the information loaded won't be the same.
GFF and BED files describe features within a region, providing a chromosome, start and end. All the variants between the start and end will be annotated with the information.
GFF : From this file format, only the third column, containing the feature is extracted and loaded with the key "feature"
This line of GFF will generate the next additionalAttributes:
| Code Block | ||
|---|---|---|
| ||
chr22 TeleGene enhancer 16053659 16063659 500 + . touch1 |
| Code Block | ||||
|---|---|---|---|---|
| ||||
VariantAnnotation = {
// ...
"additionalAttributes" : {
"myGff" : {
"attribute" : {
"feature" : "enhancer"
}
}
}
}
|
BED : From the bed format, columns name (4th), score (5th) and strand (6th) will be loaded.
This line of BED will generate the next additionalAttributes:
| Code Block | ||
|---|---|---|
| ||
chr22 16053659 16063659 Pos1 353 + 127471196 127472363 255,0,0 0 A A |
| Code Block | ||||
|---|---|---|---|---|
| ||||
VariantAnnotation = {
// ...
"additionalAttributes" : {
"myBed" : {
"attribute" : {
"name":"Pos1",
"score":"353",
"strand":"+"
}
}
}
}
|
VCF : This format is not region based, so each line will modify a single variant. All the INFO column will be loaded as additional attributes.
The next VCF will generate the next additionalAttributes:
| Code Block | ||
|---|---|---|
| ||
##fileformat=VCFv4.2
##FILTER=<ID=PASS,Description="All filters passed">
##INFO=<ID=FEATURE,Number=1,Type=String,Description="Feature type">
##INFO=<ID=SCORE,Number=1,Type=Integer,Description="Score value">
##INFO=<ID=STRAND,Number=1,Type=Integer,Description="Strand">
#CHROM POS ID REF ALT QUAL FILTER INFO
chr22 16050075 A G . 100 PASS FEATURE=specific;SCORE=300;STRAND=+ |
| Code Block | ||||
|---|---|---|---|---|
| ||||
VariantAnnotation = {
// ...
"additionalAttributes" : {
"myVcf" : {
"attribute" : {
"FEATURE":"specific",
"SCORE":"300",
"STRAND":"+"
}
}
}
}
|
| Code Block | ||||
|---|---|---|---|---|
| ||||
VariantAnnotation = {
// ...
"additionalAttributes" : {
"myVcf" : {
"attribute" : {
"FEATURE":"specific",
"SCORE":"300",
"STRAND":"+"
}
},
"myBed" : {
"attribute" : {
"name":"Pos1",
"score":"353",
"strand":"+"
}
}
}
} |
Custom Annotator
Calculate Statistics
Pre-calculated stats are useful for filtering variants.
Define cohorts
Remove
Export / Query
Export frequencies (statistics)
Export frequencies (statistics) is an special case of export. Instead of export full variants, only the variant cohort statistics are exported.
To export variant frequencies, use the command variant export-frequencies in the command line.
| Code Block |
|---|
opencga-analysis.sh variant export-frequencies -s <study> --output-format <vcf|tsv|cellbase|json>
opencga-storage.sh variant export-frequencies -s <study> --output-format <vcf|tsv|cellbase|json |
As for variants export, there are multiple possible output formats:
VCF : Standard VCF format without samples information, with the stats as values in the INFO column.
| Code Block | ||||
|---|---|---|---|---|
| ||||
##fileformat=VCFv4.2
##FILTER=<ID=.,Description="No FILTER info">
##FILTER=<ID=PASS,Description="Valid variant">
##INFO=<ID=AC,Number=A,Type=Integer,Description="Total number of alternate alleles in called genotypes, for each ALT allele, in the same order as listed">
##INFO=<ID=AF,Number=A,Type=Float,Description="Allele Frequency, for each ALT allele, calculated from AC and AN, in the range (0,1), in the same order as listed">
##INFO=<ID=AN,Number=1,Type=Integer,Description="Total number of alleles in called genotypes">
##INFO=<ID=AFK_AF,Number=A,Type=Float,Description="Allele frequency in the C1 cohort calculated from AC and AN, in the range (0,1), in the same order as listed">
#CHROM POS ID REF ALT QUAL FILTER INFO
22 16050115 . G A . PASS AC=1;AF=0.001;AN=1000;AFK_AF=0.002008
22 16050213 . C T . PASS AC=1;AF=0.001;AN=1000;AFK_AF=0
22 16050319 . C T . PASS AC=1;AF=0.001;AN=1000;AFK_AF=0
22 16050607 . G A . PASS AC=2;AF=0.002;AN=1000;AFK_AF=0.004016
|
TSV (Tab Separated Values). Simple format with each cohort in one column.
| Code Block | ||||
|---|---|---|---|---|
| ||||
#CHR POS REF ALT ALL_AN ALL_AC ALL_AF ALL_HET ALL_HOM
22 16050213 C T 1000 1 0.001 0.002 0.0
22 16050607 G A 1000 2 0.002 0.004 0.0
22 16050740 A - 1000 1 0.001 0.002 0.0
22 16050840 C G 1000 13 0.013 0.026 0.0
22 16051075 G A 1000 2 0.002 0.004 0.0
22 16051249 T C 1000 91 0.091 0.162 0.01
22 16051453 A C 998 74 0.074 0.144 0.004
22 16051453 A G 926 2 0.002 0.144 0.004
22 16051723 A - 1000 12 0.012 0.024 0.0
22 16051816 T G 1000 2 0.002 0.004 0.0
|
JSON. Variant model just with minimal information and statistics.
| Code Block | ||||
|---|---|---|---|---|
| ||||
{"reference":"G","names":[],"chromosome":"22","alternate":"A","start":16050115,"annotation":null,"id":"22:16050115:G:A","type":"SNV","studies":[{"format":[],"samplesData":[],"studyId":"user@p1:s1","stats":{"C3":{"refAllele":"G","altAllele":"A","refAlleleCount":2,"altAlleleCount":0,"genotypesCount":{"0/0":1},"genotypesFreq":{"0/0":1.0},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":1.0,"altAlleleFreq":0.0,"maf":0.0,"mgf":0.0,"mafAllele":"A","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"ALL":{"refAllele":"G","altAllele":"A","refAlleleCount":999,"altAlleleCount":1,"genotypesCount":{"0/0":499,"0|1":1},"genotypesFreq":{"0/0":0.998,"0|1":0.002},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":0.999,"altAlleleFreq":0.001,"maf":0.001,"mgf":0.0,"mafAllele":"A","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"C4":{"refAllele":"G","altAllele":"A","refAlleleCount":-1,"altAlleleCount":-1,"genotypesCount":{},"genotypesFreq":{},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":2.0,"altAlleleFreq":-1.0,"maf":-1.0,"mgf":-1.0,"mafAllele":null,"mgfGenotype":null,"passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"C1":{"refAllele":"G","altAllele":"A","refAlleleCount":500,"altAlleleCount":0,"genotypesCount":{"0/0":250},"genotypesFreq":{"0/0":1.0},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":1.0,"altAlleleFreq":0.0,"maf":0.0,"mgf":0.0,"mafAllele":"A","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"C2":{"refAllele":"G","altAllele":"A","refAlleleCount":497,"altAlleleCount":1,"genotypesCount":{"0/0":248,"0|1":1},"genotypesFreq":{"0/0":0.99598396,"0|1":0.004016064},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":0.997992,"altAlleleFreq":0.002008032,"maf":0.002008032,"mgf":0.0,"mafAllele":"A","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"}},"files":[],"secondaryAlternates":[]}],"end":16050115,"strand":"+","sv":null,"hgvs":{},"length":1}
{"reference":"C","names":[],"chromosome":"22","alternate":"T","start":16050213,"annotation":null,"id":"22:16050213:C:T","type":"SNV","studies":[{"format":[],"samplesData":[],"studyId":"user@p1:s1","stats":{"C3":{"refAllele":"C","altAllele":"T","refAlleleCount":2,"altAlleleCount":0,"genotypesCount":{"0/0":1},"genotypesFreq":{"0/0":1.0},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":1.0,"altAlleleFreq":0.0,"maf":0.0,"mgf":0.0,"mafAllele":"T","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"ALL":{"refAllele":"C","altAllele":"T","refAlleleCount":999,"altAlleleCount":1,"genotypesCount":{"0|1":1,"0/0":499},"genotypesFreq":{"0|1":0.002,"0/0":0.998},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":0.999,"altAlleleFreq":0.001,"maf":0.001,"mgf":0.0,"mafAllele":"T","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"C4":{"refAllele":"C","altAllele":"T","refAlleleCount":-1,"altAlleleCount":-1,"genotypesCount":{},"genotypesFreq":{},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":2.0,"altAlleleFreq":-1.0,"maf":-1.0,"mgf":-1.0,"mafAllele":null,"mgfGenotype":null,"passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"C1":{"refAllele":"C","altAllele":"T","refAlleleCount":500,"altAlleleCount":0,"genotypesCount":{"0/0":250},"genotypesFreq":{"0/0":1.0},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":1.0,"altAlleleFreq":0.0,"maf":0.0,"mgf":0.0,"mafAllele":"T","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"C2":{"refAllele":"C","altAllele":"T","refAlleleCount":497,"altAlleleCount":1,"genotypesCount":{"0|1":1,"0/0":248},"genotypesFreq":{"0|1":0.004016064,"0/0":0.99598396},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":0.997992,"altAlleleFreq":0.002008032,"maf":0.002008032,"mgf":0.0,"mafAllele":"T","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"}},"files":[],"secondaryAlternates":[]}],"end":16050213,"strand":"+","sv":null,"hgvs":{},"length":1}
{"reference":"C","names":[],"chromosome":"22","alternate":"T","start":16050319,"annotation":null,"id":"22:16050319:C:T","type":"SNV","studies":[{"format":[],"samplesData":[],"studyId":"user@p1:s1","stats":{"C3":{"refAllele":"C","altAllele":"T","refAlleleCount":2,"altAlleleCount":0,"genotypesCount":{"0/0":1},"genotypesFreq":{"0/0":1.0},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":1.0,"altAlleleFreq":0.0,"maf":0.0,"mgf":0.0,"mafAllele":"T","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"ALL":{"refAllele":"C","altAllele":"T","refAlleleCount":999,"altAlleleCount":1,"genotypesCount":{"0/0":499,"1|0":1},"genotypesFreq":{"0/0":0.998,"1|0":0.002},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":0.999,"altAlleleFreq":0.001,"maf":0.001,"mgf":0.0,"mafAllele":"T","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"C4":{"refAllele":"C","altAllele":"T","refAlleleCount":-1,"altAlleleCount":-1,"genotypesCount":{},"genotypesFreq":{},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":2.0,"altAlleleFreq":-1.0,"maf":-1.0,"mgf":-1.0,"mafAllele":null,"mgfGenotype":null,"passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"C1":{"refAllele":"C","altAllele":"T","refAlleleCount":500,"altAlleleCount":0,"genotypesCount":{"0/0":250},"genotypesFreq":{"0/0":1.0},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":1.0,"altAlleleFreq":0.0,"maf":0.0,"mgf":0.0,"mafAllele":"T","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"C2":{"refAllele":"C","altAllele":"T","refAlleleCount":497,"altAlleleCount":1,"genotypesCount":{"0/0":248,"1|0":1},"genotypesFreq":{"0/0":0.99598396,"1|0":0.004016064},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":0.997992,"altAlleleFreq":0.002008032,"maf":0.002008032,"mgf":0.0,"mafAllele":"T","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"}},"files":[],"secondaryAlternates":[]}],"end":16050319,"strand":"+","sv":null,"hgvs":{},"length":1}
{"reference":"G","names":[],"chromosome":"22","alternate":"A","start":16050607,"annotation":null,"id":"22:16050607:G:A","type":"SNV","studies":[{"format":[],"samplesData":[],"studyId":"user@p1:s1","stats":{"C3":{"refAllele":"G","altAllele":"A","refAlleleCount":2,"altAlleleCount":0,"genotypesCount":{"0/0":1},"genotypesFreq":{"0/0":1.0},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":1.0,"altAlleleFreq":0.0,"maf":0.0,"mgf":0.0,"mafAllele":"A","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"ALL":{"refAllele":"G","altAllele":"A","refAlleleCount":998,"altAlleleCount":2,"genotypesCount":{"0/0":498,"0|1":1,"1|0":1},"genotypesFreq":{"0/0":0.996,"0|1":0.002,"1|0":0.002},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":0.998,"altAlleleFreq":0.002,"maf":0.002,"mgf":0.0,"mafAllele":"A","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"C4":{"refAllele":"G","altAllele":"A","refAlleleCount":-1,"altAlleleCount":-1,"genotypesCount":{},"genotypesFreq":{},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":2.0,"altAlleleFreq":-1.0,"maf":-1.0,"mgf":-1.0,"mafAllele":null,"mgfGenotype":null,"passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"C1":{"refAllele":"G","altAllele":"A","refAlleleCount":499,"altAlleleCount":1,"genotypesCount":{"0/0":249,"0|1":1},"genotypesFreq":{"0/0":0.996,"0|1":0.004},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":0.998,"altAlleleFreq":0.002,"maf":0.002,"mgf":0.0,"mafAllele":"A","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"},"C2":{"refAllele":"G","altAllele":"A","refAlleleCount":497,"altAlleleCount":1,"genotypesCount":{"0/0":248,"1|0":1},"genotypesFreq":{"0/0":0.99598396,"1|0":0.004016064},"missingAlleles":0,"missingGenotypes":0,"refAlleleFreq":0.997992,"altAlleleFreq":0.002008032,"maf":0.002008032,"mgf":0.0,"mafAllele":"A","mgfGenotype":"1/1","passedFilters":false,"mendelianErrors":-1,"casesPercentDominant":-1.0,"controlsPercentDominant":-1.0,"casesPercentRecessive":-1.0,"controlsPercentRecessive":-1.0,"quality":-1.0,"numSamples":-1,"variantType":"SNV"}},"files":[],"secondaryAlternates":[]}],"end":16050607,"strand":"+","sv":null,"hgvs":{},"length":1}
|
Population Frequencies (Cellbase mode). Specific JSON format for import into Cellbase variation. It is a Variant model with VariantAnnotation with PupulationFrequencies.
| Code Block | ||||
|---|---|---|---|---|
| ||||
{"names":[],"reference":"T","chromosome":"22","alternate":"C","start":16174643,"annotation":{"populationFrequencies":[{"study":"s1","population":"ALL","refAllele":"T","altAllele":"C","refAlleleFreq":0.999,"altAlleleFreq":0.001,"refHomGenotypeFreq":0.998,"hetGenotypeFreq":0.002,"altHomGenotypeFreq":0.0},{"study":"s1","population":"C1","refAllele":"T","altAllele":"C","refAlleleFreq":0.998,"altAlleleFreq":0.002,"refHomGenotypeFreq":0.996,"hetGenotypeFreq":0.004,"altHomGenotypeFreq":0.0}]},"end":16174643,"type":"SNV","studies":[],"strand":"+","hgvs":{},"length":1}
{"names":[],"reference":"C","chromosome":"22","alternate":"T","start":16176715,"annotation":{"populationFrequencies":[{"study":"s1","population":"ALL","refAllele":"C","altAllele":"T","refAlleleFreq":0.998,"altAlleleFreq":0.002,"refHomGenotypeFreq":0.996,"hetGenotypeFreq":0.004,"altHomGenotypeFreq":0.0},{"study":"s1","population":"C2","refAllele":"C","altAllele":"T","refAlleleFreq":0.99598396,"altAlleleFreq":0.004016064,"refHomGenotypeFreq":0.99196786,"hetGenotypeFreq":0.008032128,"altHomGenotypeFreq":0.0}]},"end":16176715,"type":"SNV","studies":[],"strand":"+","hgvs":{},"length":1}
{"names":[],"reference":"C","chromosome":"22","alternate":"A","start":16176724,"annotation":{"populationFrequencies":[{"study":"s1","population":"ALL","refAllele":"C","altAllele":"A","refAlleleFreq":0.999,"altAlleleFreq":0.001,"refHomGenotypeFreq":0.998,"hetGenotypeFreq":0.002,"altHomGenotypeFreq":0.0},{"study":"s1","population":"C2","refAllele":"C","altAllele":"A","refAlleleFreq":0.997992,"altAlleleFreq":0.002008032,"refHomGenotypeFreq":0.99598396,"hetGenotypeFreq":0.004016064,"altHomGenotypeFreq":0.0}]},"end":16176724,"type":"SNV","studies":[],"strand":"+","hgvs":{},"length":1}
{"names":[],"reference":"T","chromosome":"22","alternate":"C","start":16176769,"annotation":{"populationFrequencies":[{"study":"s1","population":"ALL","refAllele":"T","altAllele":"C","refAlleleFreq":0.999,"altAlleleFreq":0.001,"refHomGenotypeFreq":0.998,"hetGenotypeFreq":0.002,"altHomGenotypeFreq":0.0},{"study":"s1","population":"C2","refAllele":"T","altAllele":"C","refAlleleFreq":0.997992,"altAlleleFreq":0.002008032,"refHomGenotypeFreq":0.99598396,"hetGenotypeFreq":0.004016064,"altHomGenotypeFreq":0.0}]},"end":16176769,"type":"SNV","studies":[],"strand":"+","hgvs":{},"length":1}
{"names":[],"reference":"T","chromosome":"22","alternate":"A","start":16176926,"annotation":{"populationFrequencies":[{"study":"s1","population":"C3","refAllele":"T","altAllele":"A","refAlleleFreq":0.5,"altAlleleFreq":0.5,"refHomGenotypeFreq":0.0,"hetGenotypeFreq":1.0,"altHomGenotypeFreq":0.0},{"study":"s1","population":"ALL","refAllele":"T","altAllele":"A","refAlleleFreq":0.473,"altAlleleFreq":0.527,"refHomGenotypeFreq":0.166,"hetGenotypeFreq":0.614,"altHomGenotypeFreq":0.22},{"study":"s1","population":"C1","refAllele":"T","altAllele":"A","refAlleleFreq":0.474,"altAlleleFreq":0.526,"refHomGenotypeFreq":0.164,"hetGenotypeFreq":0.62,"altHomGenotypeFreq":0.216},{"study":"s1","population":"C2","refAllele":"T","altAllele":"A","refAlleleFreq":0.4698795,"altAlleleFreq":0.5301205,"refHomGenotypeFreq":0.16465864,"hetGenotypeFreq":0.6104418,"altHomGenotypeFreq":0.2248996}]},"end":16176926,"type":"SNV","studies":[],"strand":"+","hgvs":{},"length":1}
|
Operations
Variant Storage Operations are responsible for leaving variant data ready for querying and analysis, for instance VCF loading, integrity checks, sample genotype aggregation, indexing, or variant annotation are examples of operations. Operations can only be executed by admin users. Many operations write and update indexed data, this will significantly improve the quality and performance of different queries and analysis.
The OpenCGA Variant Storage Engine supports several operations to work with variant datasets:
Index Pipeline
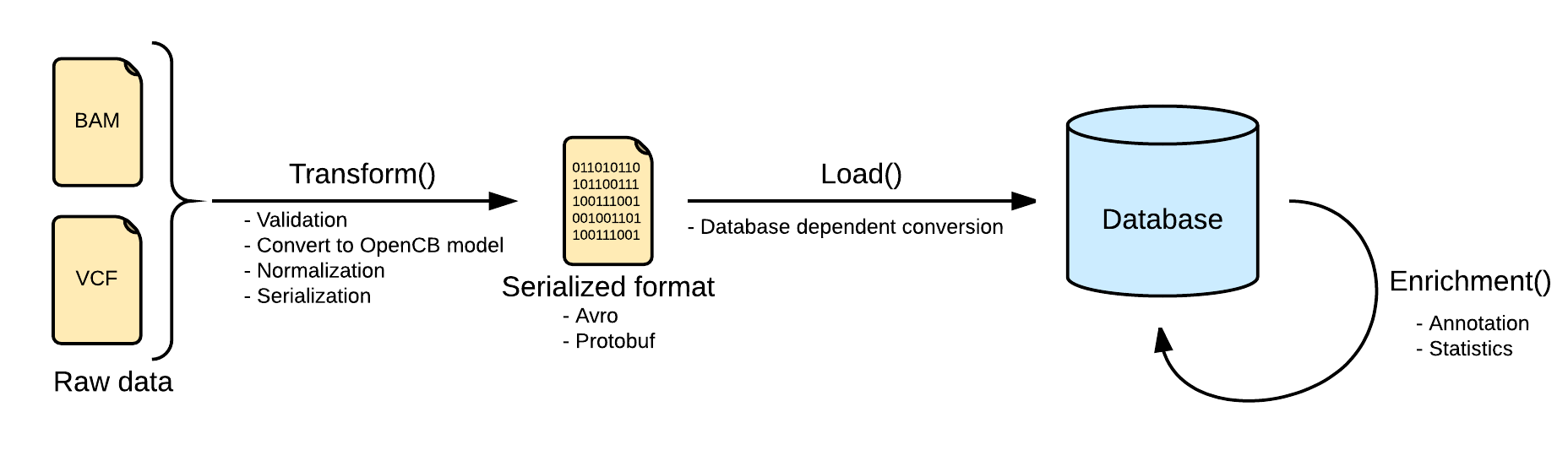 Image Added
Image Added
Table of Contents:
| Table of Contents | ||
|---|---|---|
|