Versions Compared
Key
- This line was added.
- This line was removed.
- Formatting was changed.
Overview
OpenCGA implements a Python client library called PyOpenCGA to perform any operation through the REST web services API. PyOpenCGA provides programmatic access to all the implemented REST webservices, providing and easy, lightweight, fast and intuitive solution to access OpenCGA data. The library offers the convenience of an object-oriented scripting language and provides the ability to integrate the obtained results into other Python applications.
Some of the main features include:
- full RESTful web service API implemented, all endpoints are supported including new alignment or clinical functionality.
- data is returned in a new RestResponse object which contains metadata and the results, some handy methods and iterators implemented.
- it uses the OpenCGA client-configuration.yml file.
- several Jupyter Notebooks implemented.
PyOpenCGA has been implemented and contributed by Pablo Marin and David Gomez and it is based on the previous pyCGA library implemented by Antonio Rueda and Daniel Perez from Genomics England. The code is open-source and can be found at https://github.com/opencb/opencga/tree/develop/opencga-client/src/main/python/pyOpenCGA. It can be easily installed using PyPI. Please, find more details on how to use the python library at Using the Python client.
Installation
Python client requires at least Python 3.x, although most of the code is fully compatible with Python 2.7. You can install PyOpenCGA either from PyPI repository or from the source code.
PyPI
PyOpenCGA client is deployed at PyPI and it is available at https://pypi.org/project/pyopencga/. Installing it is as simple as running the following command line:
| Code Block | ||||
|---|---|---|---|---|
| ||||
## Latest stable version pip install pyopencga |
Source Code
Python client source code can be found in OpenCGA GitHub repository at https://github.com/opencb/opencga/tree/develop/opencga-client/src/main/python/pyOpenCGA. To install any stable of development version of pyOpenCGA we will first need to clone the right branch of OpenCGA repository and install the library using the setup.py file.
| Code Block | ||||
|---|---|---|---|---|
| ||||
## Latest stable version git clone -b master https://github.com/opencb/opencga.git ## Move to the pyOpenCGA client folder cd opencga/opencga-client/src/main/python/pyOpenCGA/ ## Install the library python setup.py install |
Library Implementation
Developers only need to create an instance of the ClientConfiguration class passing it as an argument to the main OpenCGAClient class. They can optionally pass a valid token to start doing calls as an authenticated user.
| Code Block | ||||
|---|---|---|---|---|
| ||||
## Import OpenCGAClient and ClientConfiguration classes
from pyopencga.opencga_client import OpenCGAClient
from pyopencga.opencga_config import ClientConfiguration
## Creating a ClientConfiguration:
# This can be done by passing the path to the main client-configuration.yml file
config = ClientConfiguration('/opt/opencga/conf/client-configuration.yml')
# Or by creating a dictionary using the below format passing the OpenCGA host to point to
config = ClientConfiguration({
"rest": {
"host": "http://bioinfo.hpc.cam.ac.uk/opencga-demo"
}
})
## Create an instance of OpenCGAClient passing the configuration
oc = OpenCGAClient(config)
## Authenticate the user. Password is optional and if this is not passed to the login method, it will be prompted to the user
oc.login('user')
# or
oc.login('user', 'password') |
Design
OpenCGAClient class works as a factory containing all the different clients, one per resource, that are necessary to call to any REST web service.
| Code Block | ||||
|---|---|---|---|---|
| ||||
## Main clients user_client = oc.users project_client = oc.projects study_client = oc.studies file_client = oc.files sample_client = oc.samples cohort_client = oc.cohorts individual_client = oc.individuals family_client = oc.families clinical_client = oc.clinical job_client = oc.jobs panel_client = oc.panels ## Analysis clients variant_client = oc.variant alignment_client = oc.alignment ga4gh_client = oc.ga4gh ## Administrative clients meta_client = oc.meta admin_client = oc.admin |
The list of available actions that can be performed with all those clients can be checked in Swagger as explained in RESTful Web Services#Swagger. Each particular client has a method defined for each available web service implemented for the resource. For instance, the whole list of actions available for the Sample resource are shown above.
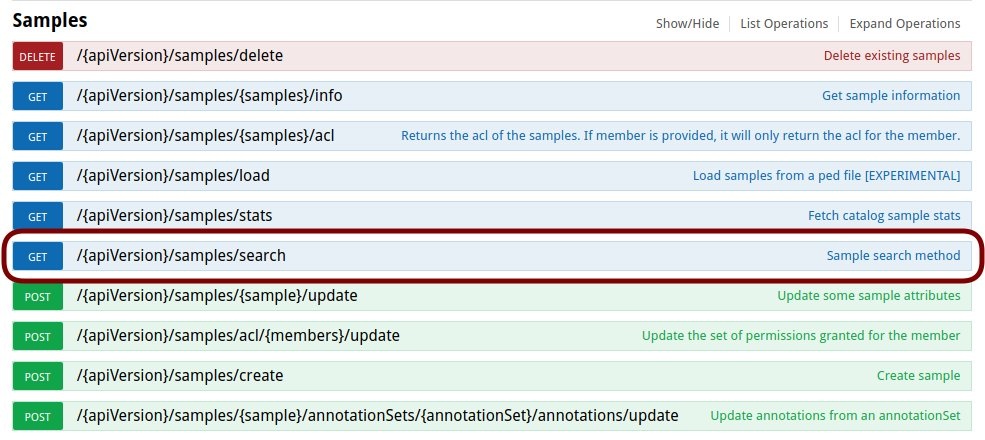
For all those actions, there is a method available in the sample client. For instance, to search for samples using the /search web service, we will need to do something like:
| Code Block | ||||
|---|---|---|---|---|
| ||||
## Look for the id of the first 5 samples of the study "study" sample_result = oc.samples.search(study="study", limit=5, include="id") |
As described in RESTful Web Services#RESTResponse, most of the web services return a RESTResponse object containing a list of OpenCGAResults. This structure has been maintained in the Python library and everytime a call to any WS is done, the response is automatically encapsulated into a custom RESTResponse class that automatically stores all the different values returned. For instance, the sample_result variable from the example above is a RESTResponse instance. This object defines a few public methods to help users navigating through the data.
The RESTResponse methods developed are:
| Code Block | ||||
|---|---|---|---|---|
| ||||
## Return an iterator to help iterating over all the results.
sample_result.results()
## Return the total number of matches taking of all the QueryResponses.
sample_result.num_matches()
## Return the total number of results taking of all the QueryResponses.
sample_result.num_results() |
Examples and tutorials
A full first example of how to use and perform some queries can be found below:
| Code Block | ||||||||
|---|---|---|---|---|---|---|---|---|
| ||||||||
# First, we need to import both the ClientConfiguration and the OpenCGAClient from pyopencga.opencga_config import ClientConfiguration from pyopencga.opencga_client import OpenCGAClient # The main client-configuration.yml file has a 'host' section to point to the Rest OpenCGA endpoints. # We need to either pass the path to the configuration file or a dictionary with the format of the file. config = ClientConfiguration('/opt/opencga/conf/client-configuration.yml') config = ClientConfiguration({ "rest": { "host": "http://bioinfo.hpc.cam.ac.uk/opencga-demo" } }) # And finally create an instance of the OpenCGAClient passing the configuration oc = OpenCGAClient(config) # Now we need to authenticate. oc.login('demo') # If done this way, password will be prompted to the user so it is not displayed but... oc.login('demo', 'demo') # ... it is also possible to pass the password directly as an additional parameter # Let's assume our installation already has been populated and we are interested in looking for # all the families containing a concrete disorder: 'Rod-cone dystrophy'. To fetch this data, we will need to: family_query_response = oc.families.search(study="study1", limit=10, disorders="Rod-cone dystrophy", include="id,members.id") # Running oc.families.search(disorders="Rod-cone dystrophy") with only the 'disorders' field would only work # if only one project and one study has been defined. However, we expect that most of the OpenCGA installations # will have more than one study, so we need to specify the families of which study we are looking for. # Additionally, we are passing limit = 10 to limit the number of family results we want to fetch. Because this # is an example, we are simply limiting the number of results to 10. # Finally, if we don't specify anything else, all the values from the Family will be fetched. When writing # scripts, we are normally interested in just a few fields of a whole entry, so adding the include/exclude fields # will definitely help us getting the results faster as we will avoid sending data we are going to discard through # the network. In this particular case, we are only interested in getting the Family id and the id of the members # of the family. To know what fields you can include/exclude, please follow the data models we have defined. # family_query_response is an instance of the QueryResponse class defined in the Python library. To read the fields, # we could do the following: family_query_response.time # Get the time spent with the REST call family_query_response.apiVersion # Get the API version of the REST family_query_response.queryOptions # Get the QueryOptions of the call (include/exclude, limit, skip, count...) family_query_response.warning # Get warning messages family_query_response.error # Get error messages family_query_response.responses # Get the responses (Array of QueryResults containing the data queried) # We can iterate over all the results to print all the id's using the 'results()' method such as in the example below: for family in family_query_response.results(): print (family['id']) # We could have this same behaviour if we run the following script, which is why 'results()' is that useful. for query_result in family_query_response.responses: for family in query_result['results']: print (family['id']) # If we want to know exactly the amount of results obtained, we can run: family_query_response.num_results() # Or let's say that instead of querying the data, we only wanted to get the number of families in the study with that disorder. In that case, we could: family_count_response = oc.families.search(study="study1", disorders="Rod-cone dystrophy", count=True) # And then get the number of matches by calling to the num_matches method: family_count_response.num_matches() # Now that we know how to work with the OpenCGA QueryResponse object, we will write a script to fetch all the variants # falling in the 'BMPR2' gene found in any member of the family. In this case, we will limit the variant query to a maximum # of 10 results excluding the sample information (sample information can be huge and would make this query much slower). for family in oc.families.search(study="study1",limit=10,disorders="Rod-cone dystrophy",include="id").results(): print ("Family: " + family['id']) variant_response = oc.variant.query(family=family['id'], gene= 'BMPR2', study='study1', includeSamples=None, limit=10) if variant_response.num_total_results() > 0: for variant in variant_response.results(): print (variant['chromosome'] + ":" + str(variant['start']) + "-" + str(variant['end']) + '\t' + variant['type']) else: print ("No variant results found") print() |
Additionally, there are several notebooks defined in https://github.com/opencb/opencga/tree/develop/opencga-client/src/main/python/pyOpenCGA/notebooks with more real examples.