Versions Compared
Key
- This line was added.
- This line was removed.
- Formatting was changed.
One of the aims of The 100,000 Genomes Project from Genomics England is to enable new medical research. Researchers will study how best to use genomics in healthcare and how best to interpret the data to help patients. The causes, diagnosis and treatment of disease will also be investigated. This is currently the largest national sequencing project of its kind in the world.
In order to provide a platform capable to provide the researchers with required functionalityresearch environment, the data was indexed using OpenCGAin OpenCGA using the BigData Hadoop Platform.
Here you can find a full report of about loading 64,078 samples for Genomics England Research environment.
Platform
The platform used for this case study consists on a Hadoop Cluster of 35 nodes (5 + 30) and a LSF queue system:
| Node | #nodes | cores | memory (GB) |
|---|---|---|---|
| LSF queue node for load | 10 | 12 | 364 |
| Hadoop master nodes | 5 | 28 | 216 |
| Hadoop worker nodes | 30 | 28 | 216 |
Data
The data of this case study contains a total of 64,078 samples divided in 4 different datasets.
| Dataset | Alias | Files | File type | Samples | Samples per file | Variants |
|---|---|---|---|---|---|---|
| Rare Disease GRCh37 | RD37 | 5,329 | Multi sample VCF | 12,142 | 2.28 | 298,763,059 |
| Rare Disease GRCh38 | RD38 | 16,591 | Multi sample VCF | 33,180 | 2.00 | 437,740,498 |
| Cancer Germline GRCh38 | CG38 | 9,167 | Single sample VCF | 9,167 | 1.00 | 286,136,051 |
| Cancer Somatic GRCh38 | CS38 | 9,589 | Somatic VCF | 9,589 | 1.00 | 398,402,166 |
| Total | 40,676 | 64,078 | 1,421,041,774 | |||
Each dataset is loaded in OpenCGA as a study, grouped in 3 different projects, depending on the type of data (germline / somatic), and assembly (GRCh37 / GRCh38).
Loading Data
The data ingestion is executed in the LSF nodes, connected directly to the Hadoop platform. This configuration allows us to run multiple files at the same time.
Having 10 worker nodes in the queue, each of them loading up to 6 files at the same time, results in 60 files being loaded concurrently.
Multi sample files
The files from Rare Disease studies (RD38 & RD37) contain more than one sample per file. In average, 2 samples per file.
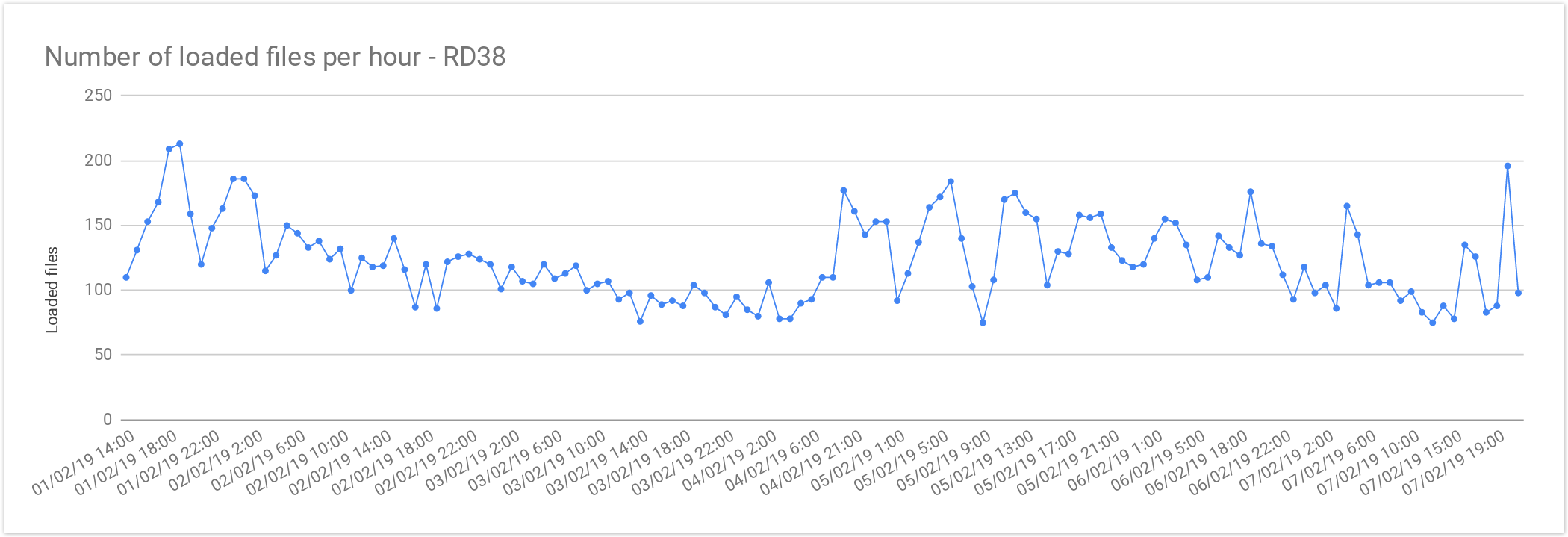
| Concurrent files loaded | 60 |
|---|---|
| Average files loaded per hour | 125.72 |
| Load time per file | 00:28:38 |
Single sample files
The files from Cancer Germline studies (CG38) contain one sample per file. Compared with the Rare Disease, these files are smaller in size, therefore, the load is slightly faster.
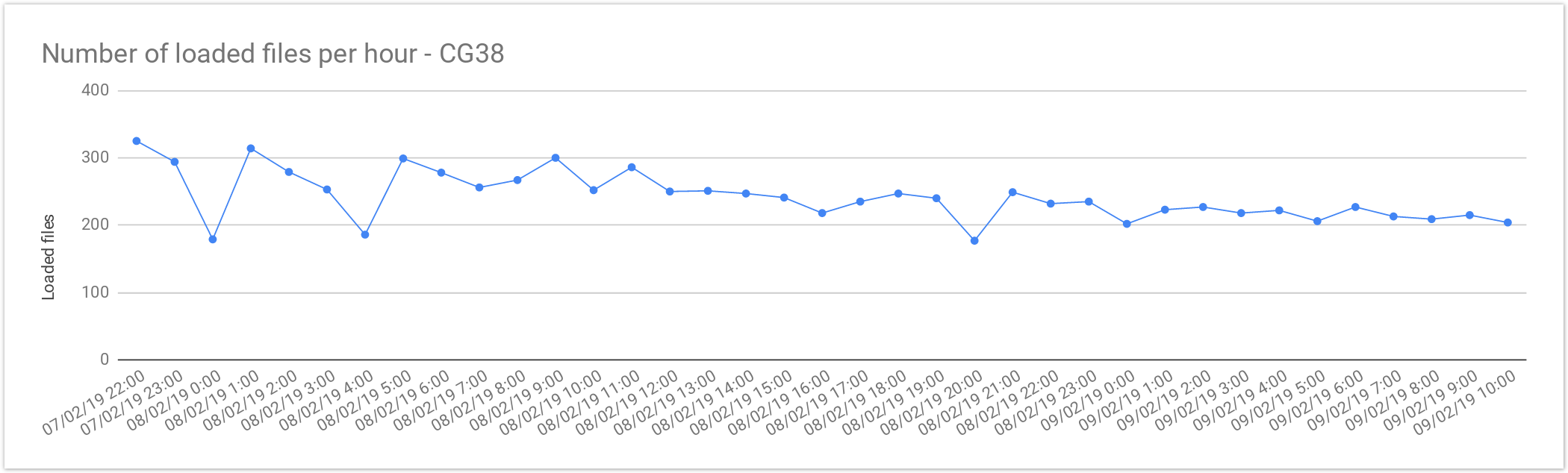
| Concurrent files loaded | 60 |
|---|---|
| Average files loaded per hour | 242.05 |
| Load time per file | 00:14:52 |
Query Performance
We would like to distinguish two types of queries: General and Clinical
General Queries
These queries are are only filtering by variant annotation and cohort stats. These queries only include aggregated data, not returning sample genotypes.
| Filter | Results | Total Results | Time |
|---|---|---|---|
| consequence type = LoF + missense_variant | 10 | 3704626 | 0.189s |
consequence type = LoF + missense_variant biotype = protein_coding | 10 | 3576472 | 0.260s |
panel with 200 genes | 10 | 3882902 | 0.299s |
Clinical Queries
Clinical queries, or sample queries, enforces queries to return variants of a specific set of samples. These queries can use all the filters from the general queries. The result will include a ReportedEvent for each variant, which determines possible conditions associated to the variant.
| Filter | Results | Total Results | Time |
|---|---|---|---|
Segregation mode = biallelic filter = PASS | 10 | 211787 | 0.420s |
Segregation mode = biallelic filter = PASS | 2000 | 211787 | 1.079s |
De novo variants filter = PASS consequence type = LoF + missense_variant | 24 | 24 | 0.680s |
| Compound Heterozygous filter = PASS biotype = protein_coding consequence type = LoF + missense_variant | 717 | 717 | 10.995s |
Table of Contents:
| Table of Contents | ||
|---|---|---|
|