Versions Compared
Key
- This line was added.
- This line was removed.
- Formatting was changed.
Pre-requisites
In order to follow this guide you have to install BioNetDB in your system. Please, please follow the steps on installation guide Installation Guide and set it up.
Download test data
Users can download test data from the following link. Download the tar.gz file and uncompress in your system. Once uncompressed, you should see the following files:
Download the test data from http://bioinfo.hpc.cam.ac.uk/downloads/bionetdb/bionetdb.dataset.tar.gz and extract the content of the archive executing:
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
# Download in the /tmp folder
$ cd /tmp
$ wget http://bioinfo.hpc.cam.ac.uk/downloads/bionetdb/bionetdb.dataset.tar.gz
# Extract the content
$ tar xvfz bionetdb.dataset.tar.gz
bionetdb.dataset/
bionetdb.dataset/illumina_platinum.export.5k.json
bionetdb.dataset/mirna.csv
bionetdb.dataset/genes.json.gz
bionetdb.dataset/proteins.json.gz
bionetdb.dataset/illumina_platinum.export.5k.json.meta.json
bionetdb.dataset/Homo_sapiens.owl
bionetdb.dataset/10k.clinvar.json.gz
# List the content
$ cd bionetdb.dataset/
$ ls -ltrh
total 475M
-rw-rw-r-- 1 jtarraga jtarraga 38M Jun 26 13:39 proteins.json.gz
-rw-rw-r-- 1 jtarraga jtarraga 78M Jun 26 13:39 genes.json.gz
-rw-rw-r-- 1 jtarraga jtarraga 1.2M Jun 26 13:39 mirna.csv
-rw-rw-r-- 1 jtarraga jtarraga 53K Jun 26 13:39 illumina_platinum.export.5k.json.meta.json
-rw-rw-r-- 1 jtarraga jtarraga 56M Jun 26 13:39 illumina_platinum.export.5k.json
-rw-rw-r-- 1 jtarraga jtarraga 215M Jun 26 13:39 Homo_sapiens.owl
-rw-rw-r-- 1 jtarraga jtarraga 89M Jun 26 13:39 10k.clinvar.json.gz |
Import genomic data
Before you query BioNetDB database, you have to populate it by importing your . Neo4j provides a mechanism to do batch imports of large amounts of data into the a Neo4j database from CSV files. BioNetDB provides a command line interface to import data. First, you prepare your data, and then, you load into the BioNetDB database:
Prepare your data, i.e., transform your genomic data files into Neo4j CSV files:| Code Block | ||
|---|---|---|
| ||
./bionetdb.sh import -i <input-directory> -o <output-csv-directory> --create-csv-files |
| title | Load CSV files |
|---|
The importing mechanism has been integrated in the BioNetDB command line (bionetdb.sh import) that allows users, first, prepare your data by creating the Neo4j CSV files, and then, these files are loaded into the database.
Creating the Neo4j CSV files
In order to create the Neo4j CSV files you have to use the BioNetDB command line: bionetdb.sh import --create-csv. The following command line creates the Neo4j CSV files for the previously downloaded dataset.
| Code Block | ||||
|---|---|---|---|---|
| ||||
$ mkdir /tmp/bionetdb.dataset/csv
$ ./bionetdb.sh import -i /tmp/bionetdb.dataset -o /tmp/bionetdb.dataset/csv --create-csv-files
...
...
[main] INFO org.opencb.bionetdb.core.utils.Neo4jBioPaxImporter - 2: 96%
[main] INFO org.opencb.bionetdb.core.utils.Neo4jBioPaxImporter - 2: 99%
[main] INFO org.opencb.bionetdb.core.utils.Neo4jBioPaxImporter - Processing /tmp/bionetdb.dataset/Homo_sapiens.owl containing 383790 BioPax elements in 11 s
[main] INFO org.opencb.bionetdb.core.utils.Neo4jBioPaxImporter - Processing 55847 nodes
[main] INFO org.opencb.bionetdb.core.utils.Neo4jBioPaxImporter - Processing 178398 relations
[main] INFO class org.opencb.bionetdb.app.cli.ImportCommandExecutor - Post-processing 778 dna nodes
[main] INFO class org.opencb.bionetdb.app.cli.ImportCommandExecutor - Post-processing 302 miRNA nodes
[main] INFO org.opencb.bionetdb.core.utils.Neo4jCsvImporter - Processing JSON file /tmp/bionetdb.dataset/10k.clinvar.json.gz
[main] INFO org.opencb.bionetdb.core.utils.Neo4jCsvImporter - Parsing 5000 variants...
[main] INFO org.opencb.bionetdb.core.utils.Neo4jCsvImporter - Parsing 10000 variants...
[main] INFO org.opencb.bionetdb.core.utils.Neo4jCsvImporter - Parsed 10000 variants from /tmp/bionetdb.dataset/10k.clinvar.json.gz. Done!!!
[main] INFO org.opencb.bionetdb.core.utils.Neo4jCsvImporter - Processing JSON file /tmp/bionetdb.dataset/illumina_platinum.export.5k.json
[main] INFO org.opencb.bionetdb.core.utils.Neo4jCsvImporter - Parsing 5000 variants...
[main] INFO org.opencb.bionetdb.core.utils.Neo4jCsvImporter - Parsed 5000 variants from /tmp/bionetdb.dataset/illumina_platinum.export.5k.json. Done!!!
[main] INFO class org.opencb.bionetdb.app.cli.ImportCommandExecutor - Gene indexing in 40 s
[main] INFO class org.opencb.bionetdb.app.cli.ImportCommandExecutor - Protein indexing in 13 s
[main] INFO class org.opencb.bionetdb.app.cli.ImportCommandExecutor - miRNA indexing in 0 s
[main] INFO class org.opencb.bionetdb.app.cli.ImportCommandExecutor - BioPAX processing in 27 s
[main] INFO class org.opencb.bionetdb.app.cli.ImportCommandExecutor - Variant processing in 19 s |
The Neo4j CSV files are located in the output folder:
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
$ ls -ltr /tmp/bionetdb.dataset/csv
total 180936
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 VARIANT_ANNOTATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 TRANSPORT.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 TRANSCRIPT_ANNOTATION_FLAG.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 REGULATION_REGION.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 PROTEIN_ANNOTATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 PHYSICAL_ENTITY.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 PANEL.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 ONTOLOGY.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 INTERACTION.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 GENE_TRAIT_ASSOCIATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 GENE_ANNOTATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 EXPRESSION.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 EXON_OVERLAP.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 DISEASE_SUBGROUP.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 DISEASE_GROUP.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 CONFIG.csv
-rw-rw-r-- 1 jtarraga jtarraga 0 Jun 26 14:14 ASSEMBLY.csv
drwxr-xr-x 2 jtarraga jtarraga 4096 Jun 26 14:15 genes.rocksdb
drwxr-xr-x 2 jtarraga jtarraga 4096 Jun 26 14:15 mirna.rocksdb
drwxr-xr-x 2 jtarraga jtarraga 4096 Jun 26 14:15 proteins.rocksdb
drwxr-xr-x 2 jtarraga jtarraga 4096 Jun 26 14:15 rocksdb
-rw-rw-r-- 1 jtarraga jtarraga 14011261 Jun 26 14:15 XREF___PROTEIN___XREF.csv
-rw-rw-r-- 1 jtarraga jtarraga 28263017 Jun 26 14:15 XREF.csv
-rw-rw-r-- 1 jtarraga jtarraga 240044 Jun 26 14:15 VARIANT__VARIANT_CALL.csv
-rw-rw-r-- 1 jtarraga jtarraga 419286 Jun 26 14:15 VARIANT__TRAIT_ASSOCIATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 3633480 Jun 26 14:15 VARIANT__POPULATION_FREQUENCY.csv
-rw-rw-r-- 1 jtarraga jtarraga 80045 Jun 26 14:15 VARIANT_FILE_INFO__FILE.csv
-rw-rw-r-- 1 jtarraga jtarraga 516837 Jun 26 14:15 VARIANT_FILE_INFO.csv
-rw-rw-r-- 1 jtarraga jtarraga 911253 Jun 26 14:15 VARIANT.csv
-rw-rw-r-- 1 jtarraga jtarraga 793845 Jun 26 14:15 VARIANT__CONSERVATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 2068937 Jun 26 14:15 VARIANT__CONSEQUENCE_TYPE.csv
-rw-rw-r-- 1 jtarraga jtarraga 390033 Jun 26 14:15 VARIANT_CALL.csv
-rw-rw-r-- 1 jtarraga jtarraga 75421 Jun 26 14:15 UNDEFINED.csv
-rw-rw-r-- 1 jtarraga jtarraga 849048 Jun 26 14:15 TRANSCRIPT__TFBS.csv
-rw-rw-r-- 1 jtarraga jtarraga 84839 Jun 26 14:15 TRANSCRIPT__PROTEIN.csv
-rw-rw-r-- 1 jtarraga jtarraga 739714 Jun 26 14:15 TRANSCRIPT.csv
-rw-rw-r-- 1 jtarraga jtarraga 3312486 Jun 26 14:15 TFBS.csv
-rw-rw-r-- 1 jtarraga jtarraga 826 Jun 26 14:15 TARGET_GENE___MIRNA___GENE.csv
-rw-rw-r-- 1 jtarraga jtarraga 876916 Jun 26 14:15 SUBSTITUTION_SCORE.csv
-rw-rw-r-- 1 jtarraga jtarraga 1212 Jun 26 14:15 SO.csv
-rw-rw-r-- 1 jtarraga jtarraga 130465 Jun 26 14:15 SMALL_MOLECULE.csv
-rw-rw-r-- 1 jtarraga jtarraga 8913 Jun 26 14:15 RNA.csv
-rw-rw-r-- 1 jtarraga jtarraga 10839 Jun 26 14:15 REACTANT___REACTION___UNDEFINED.csv
-rw-rw-r-- 1 jtarraga jtarraga 1708 Jun 26 14:15 REACTANT___REACTION___RNA.csv
-rw-rw-r-- 1 jtarraga jtarraga 82014 Jun 26 14:15 REACTANT___REACTION___PROTEIN.csv
-rw-rw-r-- 1 jtarraga jtarraga 13002 Jun 26 14:15 REACTANT___REACTION___DNA.csv
-rw-rw-r-- 1 jtarraga jtarraga 93777 Jun 26 14:15 REACTANT___REACTION___COMPLEX.csv
-rw-rw-r-- 1 jtarraga jtarraga 39 Jun 26 14:15 PROTEIN_VARIANT_ANNOTATION__PROTEIN_KEYWORD.csv
-rw-rw-r-- 1 jtarraga jtarraga 48 Jun 26 14:15 PROTEIN_VARIANT_ANNOTATION__PROTEIN_FEATURE.csv
-rw-rw-r-- 1 jtarraga jtarraga 1558786 Jun 26 14:15 PROTEIN__PROTEIN_KEYWORD.csv
-rw-rw-r-- 1 jtarraga jtarraga 6322047 Jun 26 14:15 PROTEIN__PROTEIN_FEATURE.csv
-rw-rw-r-- 1 jtarraga jtarraga 23573 Jun 26 14:15 PROTEIN_KEYWORD.csv
-rw-rw-r-- 1 jtarraga jtarraga 74644290 Jun 26 14:15 PROTEIN_FEATURE.csv
-rw-rw-r-- 1 jtarraga jtarraga 2145473 Jun 26 14:15 PROTEIN.csv
-rw-rw-r-- 1 jtarraga jtarraga 6881 Jun 26 14:15 PRODUCT___REACTION___UNDEFINED.csv
-rw-rw-r-- 1 jtarraga jtarraga 97187 Jun 26 14:15 PRODUCT___REACTION___SMALL_MOLECULE.csv
-rw-rw-r-- 1 jtarraga jtarraga 1420 Jun 26 14:15 PRODUCT___REACTION___RNA.csv
-rw-rw-r-- 1 jtarraga jtarraga 102232 Jun 26 14:15 PRODUCT___REACTION___COMPLEX.csv
-rw-rw-r-- 1 jtarraga jtarraga 12258089 Jun 26 14:15 POPULATION_FREQUENCY.csv
-rw-rw-r-- 1 jtarraga jtarraga 15956 Jun 26 14:15 PATHWAY_NEXT_STEP___REGULATION___REGULATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 11308 Jun 26 14:15 PATHWAY_NEXT_STEP___REGULATION___CATALYSIS.csv
-rw-rw-r-- 1 jtarraga jtarraga 33343 Jun 26 14:15 PATHWAY_NEXT_STEP___REACTION___REGULATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 162451 Jun 26 14:15 PATHWAY_NEXT_STEP___REACTION___REACTION.csv
-rw-rw-r-- 1 jtarraga jtarraga 963 Jun 26 14:15 PATHWAY_NEXT_STEP___REACTION___PATHWAY.csv
-rw-rw-r-- 1 jtarraga jtarraga 127 Jun 26 14:15 PATHWAY_NEXT_STEP___PATHWAY___REGULATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 1996 Jun 26 14:15 PATHWAY_NEXT_STEP___PATHWAY___PATHWAY.csv
-rw-rw-r-- 1 jtarraga jtarraga 594 Jun 26 14:15 PATHWAY_NEXT_STEP___CATALYSIS___PATHWAY.csv
-rw-rw-r-- 1 jtarraga jtarraga 46186 Jun 26 14:15 PATHWAY_NEXT_STEP___CATALYSIS___CATALYSIS.csv
-rw-rw-r-- 1 jtarraga jtarraga 129897 Jun 26 14:15 PATHWAY.csv
-rw-rw-r-- 1 jtarraga jtarraga 34 Jun 26 14:15 MIRNA__TARGET_TRANSCRIPT.csv
-rw-rw-r-- 1 jtarraga jtarraga 968 Jun 26 14:15 MIRNA.csv
-rw-rw-r-- 1 jtarraga jtarraga 469 Jun 26 14:15 IS___RNA___MIRNA.csv
-rw-rw-r-- 1 jtarraga jtarraga 10413 Jun 26 14:15 IS___DNA___GENE.csv
-rw-rw-r-- 1 jtarraga jtarraga 94312 Jun 26 14:15 GENE__TRANSCRIPT.csv
-rw-rw-r-- 1 jtarraga jtarraga 51330 Jun 26 14:15 GENE__DRUG.csv
-rw-rw-r-- 1 jtarraga jtarraga 988165 Jun 26 14:15 GENE__DISEASE.csv
-rw-rw-r-- 1 jtarraga jtarraga 105517 Jun 26 14:15 GENE.csv
-rw-rw-r-- 1 jtarraga jtarraga 203 Jun 26 14:15 FILE.csv
-rw-rw-r-- 1 jtarraga jtarraga 120908 Jun 26 14:15 DRUG.csv
-rw-rw-r-- 1 jtarraga jtarraga 911319 Jun 26 14:15 DISEASE.csv
-rw-rw-r-- 1 jtarraga jtarraga 205 Jun 26 14:15 CONTROLLER___REGULATION___UNDEFINED.csv
-rw-rw-r-- 1 jtarraga jtarraga 67 Jun 26 14:15 CONTROLLER___REGULATION___RNA.csv
-rw-rw-r-- 1 jtarraga jtarraga 3529 Jun 26 14:15 CONTROLLER___CATALYSIS___UNDEFINED.csv
-rw-rw-r-- 1 jtarraga jtarraga 29158 Jun 26 14:15 CONTROLLER___CATALYSIS___PROTEIN.csv
-rw-rw-r-- 1 jtarraga jtarraga 41431 Jun 26 14:15 CONTROLLER___CATALYSIS___COMPLEX.csv
-rw-rw-r-- 1 jtarraga jtarraga 24238 Jun 26 14:15 CONTROLLED___REGULATION___REACTION.csv
-rw-rw-r-- 1 jtarraga jtarraga 208268 Jun 26 14:15 CONSEQUENCE_TYPE__TRANSCRIPT.csv
-rw-rw-r-- 1 jtarraga jtarraga 512125 Jun 26 14:15 CONSEQUENCE_TYPE__PROTEIN_VARIANT_ANNOTATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 38 Jun 26 14:15 CONSEQUENCE_TYPE__GENE.csv
-rw-rw-r-- 1 jtarraga jtarraga 175344 Jun 26 14:15 COMPONENT_OF_PATHWAY___REACTION___PATHWAY.csv
-rw-rw-r-- 1 jtarraga jtarraga 32800 Jun 26 14:15 COMPONENT_OF_PATHWAY___PATHWAY___PATHWAY.csv
-rw-rw-r-- 1 jtarraga jtarraga 17555 Jun 26 14:15 COMPONENT_OF_COMPLEX___UNDEFINED___COMPLEX.csv
-rw-rw-r-- 1 jtarraga jtarraga 3298 Jun 26 14:15 COMPONENT_OF_COMPLEX___RNA___COMPLEX.csv
-rw-rw-r-- 1 jtarraga jtarraga 236226 Jun 26 14:15 COMPONENT_OF_COMPLEX___PROTEIN___COMPLEX.csv
-rw-rw-r-- 1 jtarraga jtarraga 6506 Jun 26 14:15 COMPONENT_OF_COMPLEX___DNA___COMPLEX.csv
-rw-rw-r-- 1 jtarraga jtarraga 17216 Jun 26 14:15 CELLULAR_LOCATION___UNDEFINED___CELLULAR_LOCATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 44423 Jun 26 14:15 CELLULAR_LOCATION___SMALL_MOLECULE___CELLULAR_LOCATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 3384 Jun 26 14:15 CELLULAR_LOCATION___RNA___CELLULAR_LOCATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 8757 Jun 26 14:15 CELLULAR_LOCATION___DNA___CELLULAR_LOCATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 4396 Jun 26 14:15 CELLULAR_LOCATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 156427 Jun 26 14:15 CELLULAR_LOCATION___COMPLEX___CELLULAR_LOCATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 22842 Jun 26 14:15 CELLULAR_LOCATION___CATALYSIS___CELLULAR_LOCATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 117673 Jun 26 14:15 CATALYSIS.csv
-rw-rw-r-- 1 jtarraga jtarraga 33 Jun 26 14:15 XREF___RNA___XREF.csv
-rw-rw-r-- 1 jtarraga jtarraga 435530 Jun 26 14:15 VARIANT__FUNCTIONAL_SCORE.csv
-rw-rw-r-- 1 jtarraga jtarraga 240052 Jun 26 14:15 VARIANT_CALL__VARIANT_FILE_INFO.csv
-rw-rw-r-- 1 jtarraga jtarraga 2779326 Jun 26 14:15 TRAIT_ASSOCIATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 39 Jun 26 14:15 TARGET_TRANSCRIPT__TRANSCRIPT.csv
-rw-rw-r-- 1 jtarraga jtarraga 31 Jun 26 14:15 TARGET_TRANSCRIPT.csv
-rw-rw-r-- 1 jtarraga jtarraga 240043 Jun 26 14:15 SAMPLE__VARIANT_CALL.csv
-rw-rw-r-- 1 jtarraga jtarraga 97 Jun 26 14:15 SAMPLE.csv
-rw-rw-r-- 1 jtarraga jtarraga 204330 Jun 26 14:15 REGULATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 887011 Jun 26 14:15 REACTION.csv
-rw-rw-r-- 1 jtarraga jtarraga 110854 Jun 26 14:15 REACTANT___REACTION___SMALL_MOLECULE.csv
-rw-rw-r-- 1 jtarraga jtarraga 635263 Jun 26 14:15 PROTEIN_VARIANT_ANNOTATION__SUBSTITUTION_SCORE.csv
-rw-rw-r-- 1 jtarraga jtarraga 187645 Jun 26 14:15 PROTEIN_VARIANT_ANNOTATION__PROTEIN.csv
-rw-rw-r-- 1 jtarraga jtarraga 445975 Jun 26 14:15 PROTEIN_VARIANT_ANNOTATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 43355 Jun 26 14:15 PRODUCT___REACTION___PROTEIN.csv
-rw-rw-r-- 1 jtarraga jtarraga 29473 Jun 26 14:15 PATHWAY_NEXT_STEP___REGULATION___REACTION.csv
-rw-rw-r-- 1 jtarraga jtarraga 553 Jun 26 14:15 PATHWAY_NEXT_STEP___REGULATION___PATHWAY.csv
-rw-rw-r-- 1 jtarraga jtarraga 75303 Jun 26 14:15 PATHWAY_NEXT_STEP___REACTION___CATALYSIS.csv
-rw-rw-r-- 1 jtarraga jtarraga 538 Jun 26 14:15 PATHWAY_NEXT_STEP___PATHWAY___REACTION.csv
-rw-rw-r-- 1 jtarraga jtarraga 322 Jun 26 14:15 PATHWAY_NEXT_STEP___PATHWAY___CATALYSIS.csv
-rw-rw-r-- 1 jtarraga jtarraga 7221 Jun 26 14:15 PATHWAY_NEXT_STEP___CATALYSIS___REGULATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 78151 Jun 26 14:15 PATHWAY_NEXT_STEP___CATALYSIS___REACTION.csv
-rw-rw-r-- 1 jtarraga jtarraga 1366995 Jun 26 14:15 FUNCTIONAL_SCORE.csv
-rw-rw-r-- 1 jtarraga jtarraga 20391 Jun 26 14:15 DNA.csv
-rw-rw-r-- 1 jtarraga jtarraga 3052 Jun 26 14:15 CONTROLLER___REGULATION___SMALL_MOLECULE.csv
-rw-rw-r-- 1 jtarraga jtarraga 7105 Jun 26 14:15 CONTROLLER___REGULATION___PROTEIN.csv
-rw-rw-r-- 1 jtarraga jtarraga 13837 Jun 26 14:15 CONTROLLER___REGULATION___COMPLEX.csv
-rw-rw-r-- 1 jtarraga jtarraga 131 Jun 26 14:15 CONTROLLED___REGULATION___PATHWAY.csv
-rw-rw-r-- 1 jtarraga jtarraga 45 Jun 26 14:15 CONTROLLED___REGULATION___CATALYSIS.csv
-rw-rw-r-- 1 jtarraga jtarraga 73180 Jun 26 14:15 CONTROLLED___CATALYSIS___REACTION.csv
-rw-rw-r-- 1 jtarraga jtarraga 2225052 Jun 26 14:15 CONSERVATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 2521876 Jun 26 14:15 CONSEQUENCE_TYPE__SO.csv
-rw-rw-r-- 1 jtarraga jtarraga 12201659 Jun 26 14:15 CONSEQUENCE_TYPE.csv
-rw-rw-r-- 1 jtarraga jtarraga 29393 Jun 26 14:15 COMPONENT_OF_COMPLEX___SMALL_MOLECULE___COMPLEX.csv
-rw-rw-r-- 1 jtarraga jtarraga 109239 Jun 26 14:15 COMPONENT_OF_COMPLEX___COMPLEX___COMPLEX.csv
-rw-rw-r-- 1 jtarraga jtarraga 568684 Jun 26 14:15 COMPLEX.csv
-rw-rw-r-- 1 jtarraga jtarraga 5644 Jun 26 14:15 CELLULAR_LOCATION___REGULATION___CELLULAR_LOCATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 68316 Jun 26 14:15 CELLULAR_LOCATION___REACTION___CELLULAR_LOCATION.csv
-rw-rw-r-- 1 jtarraga jtarraga 244996 Jun 26 14:15 CELLULAR_LOCATION___PROTEIN___CELLULAR_LOCATION.csv |
Load Neo4j CSV files
Once created the CSV files, they have to be loaded into the database by using the BioNetDB command line: bionetdb.sh import. This command line can only be used to load data into a previously unused database, so if you are using the default Neo4j database (located at $NEO4J_HOME/data/databases/graph.db), be sure that it is empty.
According to our example:
| Code Block | ||||
|---|---|---|---|---|
| ||||
$ rm $NEO4J_HOME/data/databases/graph.db
$ ./bionetdb.sh import -i /tmp/bionetdb.dataset/csv
...
...
[>:23.27 MB/s----------|NODE:22.89 MB|*PROPERTIES(3)================|LA|v:63.93 MB/s(2)=======]2.11M ∆ 764K
Done in 6s 661ms
Prepare node index, started 2018-06-26 13:31:53.186+0000
[*DETECT:30.96 MB-----------------------------------------------------------------------------]2.12M ∆2.12M
Done in 974ms
Relationships, started 2018-06-26 13:31:54.217+0000
[*>:18.40 MB/s----------------------------------------|T|PREPARE(3)==============|RE|P|v:43.21]2.60M ∆ 376K
Done in 2s 665ms
Node Degrees, started 2018-06-26 13:31:56.955+0000
[*>(3)==========================================|CALCULATE(2)=================================]2.60M ∆2.60M
Done in 326ms
Relationship --> Relationship 1-32/32, started 2018-06-26 13:31:57.324+0000
[*>---------------------------------|LINK(4)=======================|v:??----------------------]2.60M ∆2.60M
Done in 499ms
RelationshipGroup 1-32/32, started 2018-06-26 13:31:57.844+0000
[*>:??---------------------------------------------------------------|v:??--------------------]68.6K ∆68.6K
Done in 69ms
Node --> Relationship, started 2018-06-26 13:31:57.924+0000
[>:??---|>-----------------------------------|LINK|*v:??(2)===================================]2.09M ∆2.09M
Done in 285ms
Relationship --> Relationship 1-32/32, started 2018-06-26 13:31:58.244+0000
[>-----------------------------|*LINK(2)=============================|v:??(2)=================]2.60M ∆2.44M
Done in 402ms
Count groups, started 2018-06-26 13:31:58.681+0000
[*>--------------------------------------------------------------------------------|COUNT-----]67.3K ∆67.3K
Done in 53ms
Gather, started 2018-06-26 13:31:58.804+0000
[>-------------|*CACHE------------------------------------------------------------------------]67.3K ∆67.3K
Done in 67ms
Write, started 2018-06-26 13:31:58.900+0000
[>:??---------------------------------|ENCODE----|*v:??---------------------------------------]67.0K ∆67.0K
Done in 34ms
Node --> Group, started 2018-06-26 13:31:58.957+0000
[>------------|FIRST------------------|*v:??--------------------------------------------------]14.1K ∆14.1K
Done in 21ms
Node counts, started 2018-06-26 13:31:59.012+0000
[>--------------------------------------------|*COUNT:76.29 MB--------------------------------]2.12M ∆2.12M
Done in 191ms
Relationship counts, started 2018-06-26 13:31:59.224+0000
[>(2)========================================|*COUNT(2)=======================================]2.61M ∆2.61M
Done in 256ms
IMPORT DONE in 13s 446ms.
Imported:
2117124 nodes
2605206 relationships
15047626 properties
Peak memory usage: 536.43 MB |
Accesing BioNetDB from Neo4j browser interface
You can access to your BioNetDB database from the Neo4j browser interface. Open your regular internet browser and type http://localhost:7474:
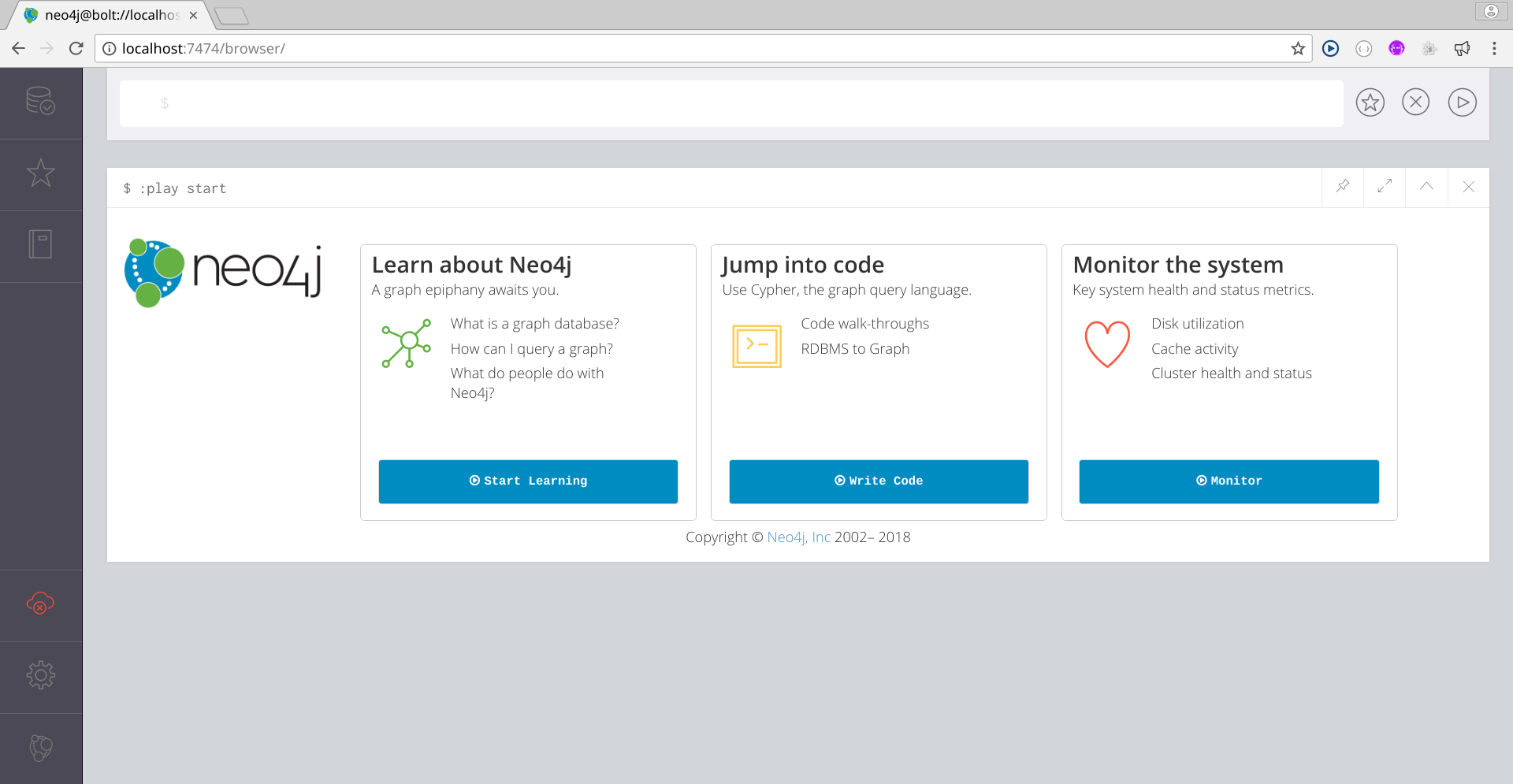 Image Added
Image Added
Now that you can access the BioNetDB database, you can start working with your imported data using the Cypher query language. For a Cypher tutorial, please refer to Intro to Cypher by the Neo4j Team.
Below you have some Cypher queries:
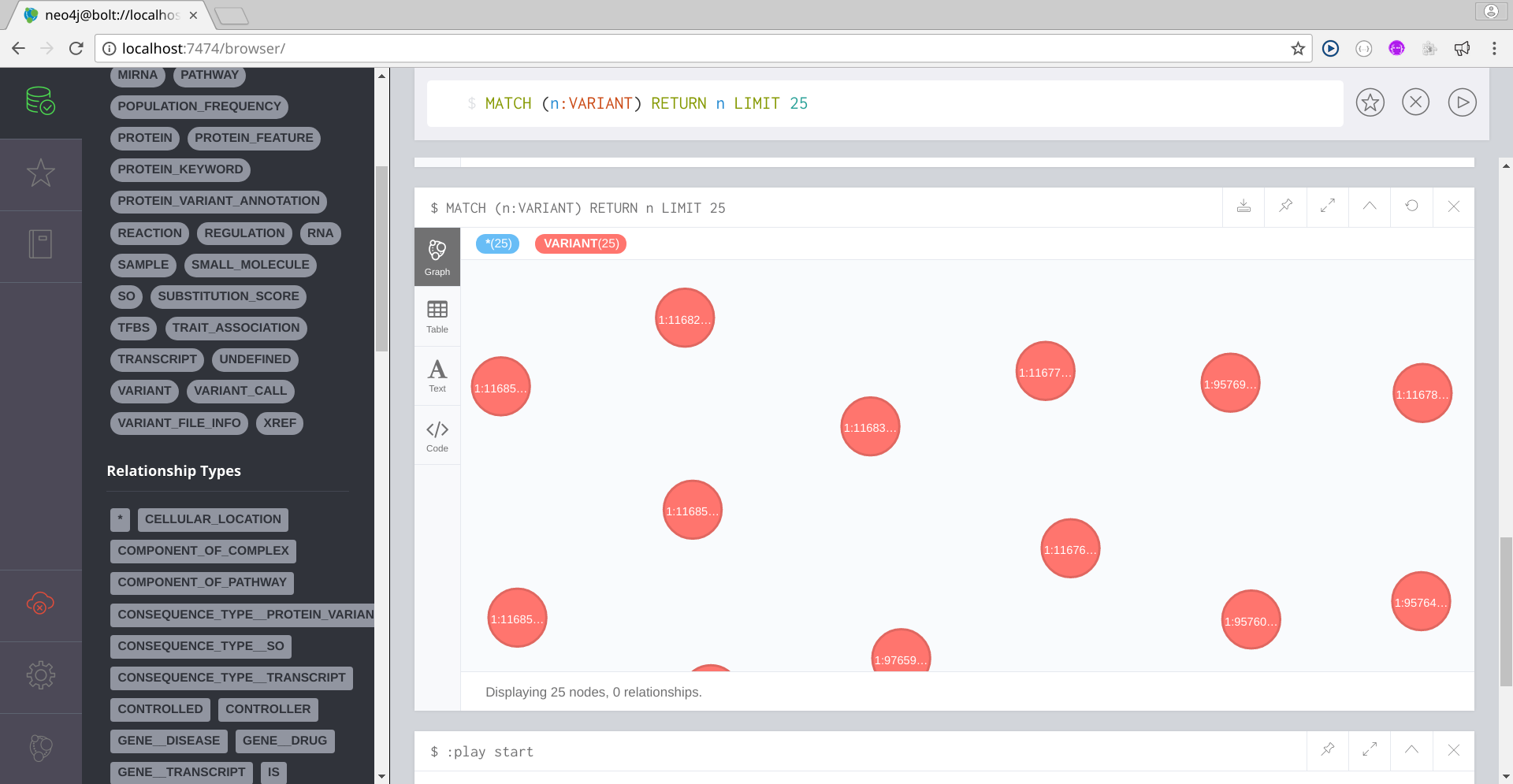 Image Added
Image Added
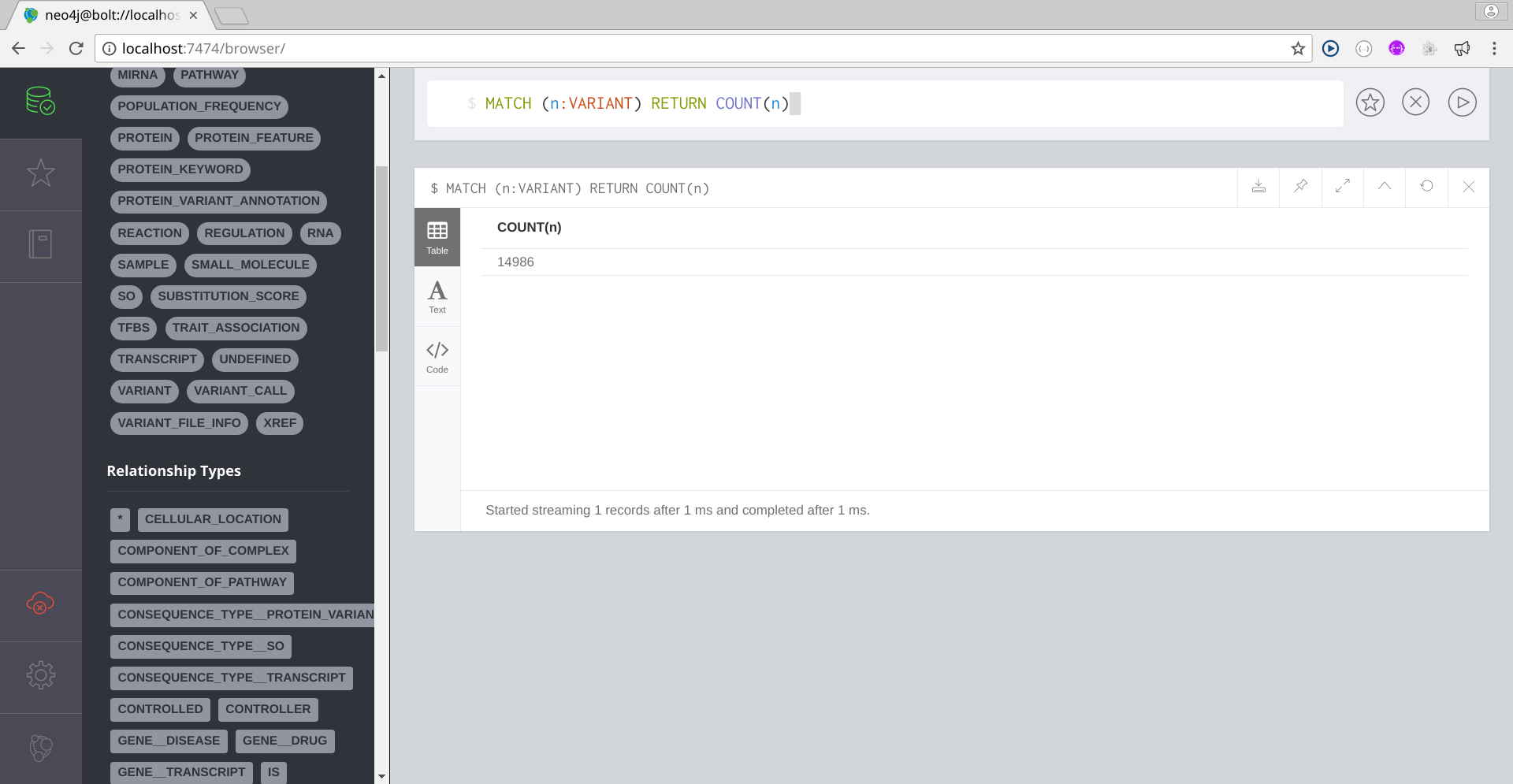 Image Added
Image Added
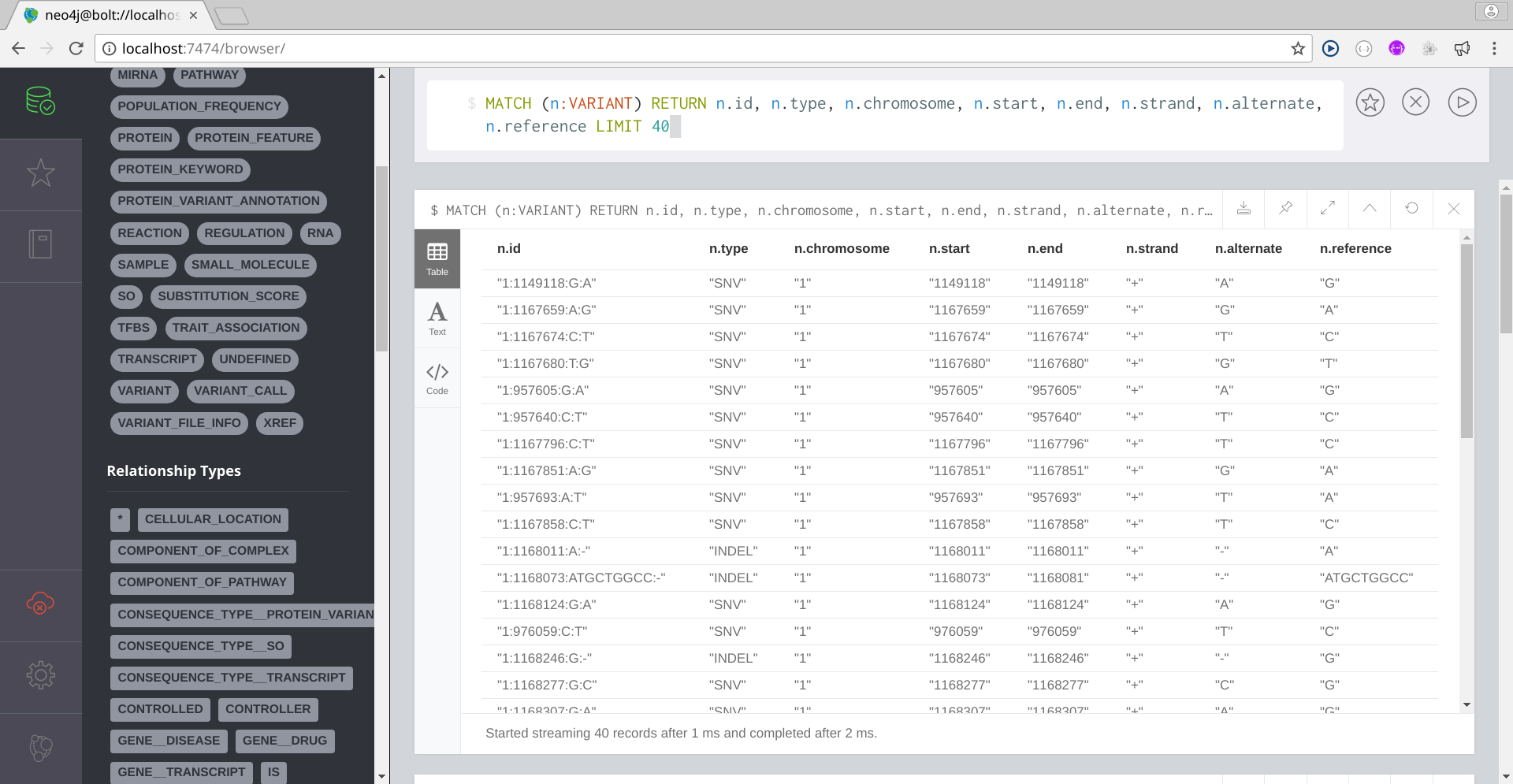 Image Added
Image Added
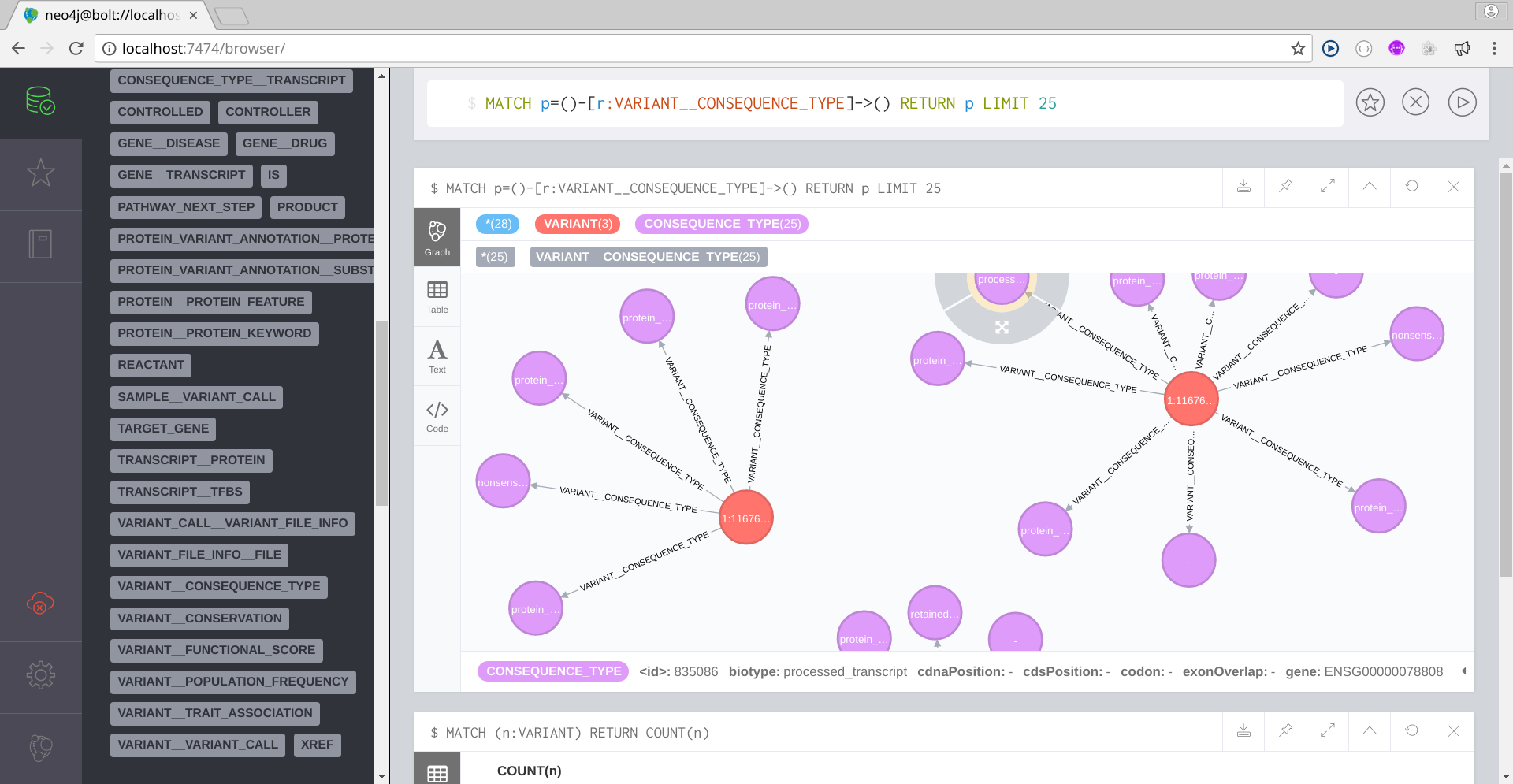 Image Added
Image Added
Table of Contents:
| Table of Contents | ||
|---|---|---|
|