...
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
field_name[value1,value2,value3...]:limit |
...
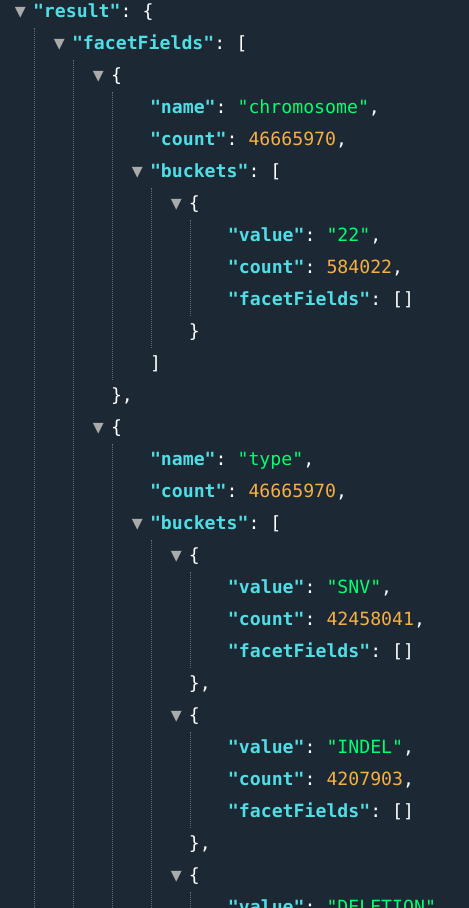
E.g.: ...&fields=chromosome[1,2];types
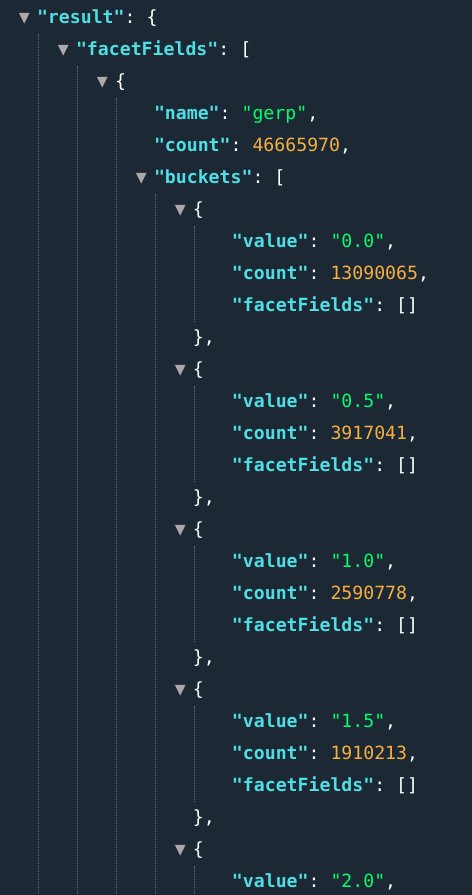
Ranges
When asking for ranges, the result contains multiple buckets over a numeric field. You must specify the field name, the lower and upper bounds and the step or bucket size.
...
E.g.: ...&fields=gerp[0..10]:0.5
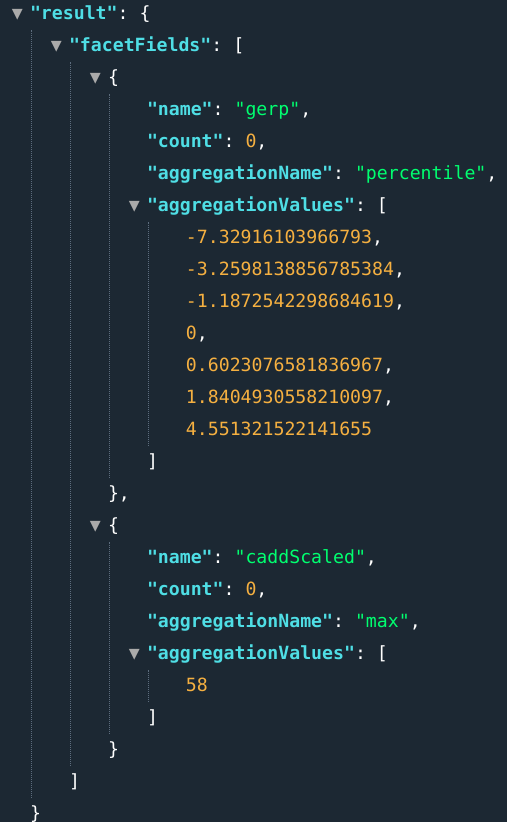
Aggregation functions
Aggregation functions, also called facet functions, analytic functions, or metrics, calculate something interesting over a domain (each facet bucket).
...
E.g.: ...&fields=percentile(gerp);max(caddScaled)
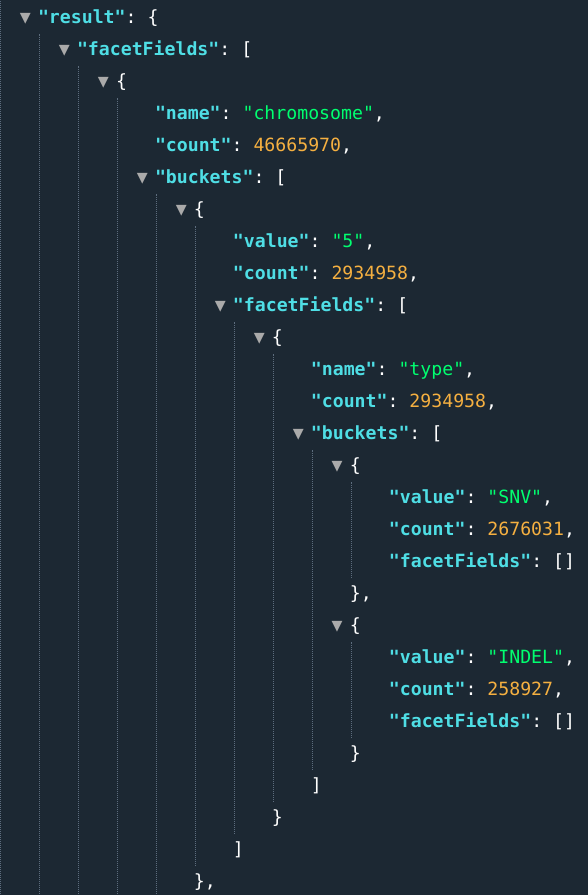
Nested facets
Nested facets allow users to nest bucketing terms, ranges or aggregations. In order to specify nested facets you must use the symbols >>
E.g.: ...&fields=chromosome[5,6]>>type
Field types: categorical and numeric
Categorical field names
| Field name | Description | Facet example |
|---|---|---|
| id | Variant ID | List of values |
| names | Names | List of <chromosome>:<start>-<end> |
| chromosome | ||
| reference | ||
| alternate | ||
| strand | ||
| type | Variant type, e.g.: INDEL, SNV, SNP,... | List of values. Accepted values: [SNV, MNV, INDEL, SV, CNV] |
| reference | Reference | List of values |
| alternate | Allternate | List of values |
| study | Matches with variants that are in the specified studies | List of values. Accept negations. |
| file | ||
| genotype | Samples with a specific genotype e.g. HG0097:0/0;HG0098:0/1,1/1. Genotype aliases accepted: HOM_REF, HOM_ALT, HET, HET_REF, HET_ALT and MISS e.g. HG0097:HOM_REF;HG0098:HET_REF,HOM_ALT. This will automatically set 'includeSample' parameter when not provided | {samp_1}:{gt_1}(,{gt_n})*(;{samp_n}:{gt_1}(,{gt_n})*)* |
| sample | Filter variants where ALL the provided samples are mutated (HET or HOM_ALT) | List of samples. |
| filter | Specify the FILTER for any of the files. If "files" filter is provided, will match the file and the filter. | List of values. |
| qual | Specify the QUAL for any of the files. If 'file' filter is provided, will match the file and the qual | |
| info | Filter by INFO attributes from file. If no file is specified, will use all files from "file" filter. e.g. AN>200 or file_1.vcf:AN>200;file_2.vcf:AN<10 . Many INFO fields can be combined. e.g. file_1.vcf:AN>200;DB=true;file_2.vcf:AN<10 | [{file}:]{key}{op}{value}[,;]* |
| format | Filter by any FORMAT field from samples. If no sample is specified, will use all samples from "sample" or "genotype" filter. e.g. DP>200 or HG0097:DP>200,HG0098:DP<10 . Many FORMAT fields can be combined. e.g. HG0097:DP>200;GT=1/1,0/1,HG0098:DP<10 | [{sample}:]{key}{op}{value}[,;]* |
| release | Return variants that were present in that specific release | Release number |
| geneTraitId | Gene trait association ID | umls:C0007222 , OMIM:269600 |
| geneTraitName | Gene trait association name | Cardiovascular Diseases |
| clinVarTrait | ClinVar trait name | |
| gwasTrait | GWAS trait name | |
| hpo | List of HPO terms. | HP:0000545 |
| go | List of GO (Genome Ontology) terms. | GO:0002020,GO:0006508 |
| expression | List of tissues of interest | |
| proteinKeywords | List of protein variant annotation keywords | |
| drug | List of drug names |
Numeric field names
Numeric field names can be used to compute range facets and aggregation functions.
| Field name | Description | Facet range example |
|---|---|---|
| start | List of genes | |
| end | ||
| length | Consequence type SO term list. | SO:0000045,SO:0000046 |
| xref | External references | |
| biotype | List of biotypes | |
| polyphen | polyphen protein substitution score | polyphen>0.1 , sift=tolerant |
| sift | sift protein substitution score | phastCons>0.5 , phylop<0.1 , gerp>0.1 |
| phastCons | phastCons conservation score | |
| phylop | phylop conservation score | |
| gerp | gerp conservation score | |
| cadd_raw | Raw CADD functional score | |
| cadd_scaled | Scaled CADD functional score | |
| maf | Minor allele frequency | |
| populationFrequencyAlt | Alternate Population Frequency | 1000GENOMES_phase_3:AFR>0.2 |
| populationFrequencyRef | Reference Population Frequency | ESP_6500:AA<0.2 |
| populationFrequencyMaf | Population minor allele frequency | EXAC:AES>=0.6 |
| transcriptionFlags | List of transcript annotation flags | CCDS, basic, cds_end_NF, mRNA_end_NF, cds_start_NF, mRNA_start_NF, seleno |
| geneTraitId | List of gene trait association ids | umls:C0007222 , OMIM:269600 |
| geneTraitName | List of gene trait association names | Cardiovascular Diseases |
| trait | List of traits, based on ClinVar, HPO, COSMIC | |
| hpo | List of HPO terms. | HP:0000545 |
| go | List of GO (Genome Ontology) terms. | GO:0002020,GO:0006508 |
| expression | List of tissues of interest | |
| proteinKeywords | List of protein variant annotation keywords | |
| drug | List of drug names |
Variant Fields
The parameters include and exclude accepts a list of Variant Fields. This is a list with all the accepted values. Some short alias to those fields are listed in italic.
- id
- chromosome
- start
- end
- reference
- alternate
- length
- type
- studies
studies.samplesData | samples | samplesData
studies.files | files
studies.stats | stats
studies.secondaryAlternates
studies.studyId
- annotation
- annotation.ancestralAllele
- annotation.id
- annotation.xrefs
- annotation.hgvs
- annotation.displayConsequenceType
- annotation.consequenceTypes
- annotation.populationFrequencies
- annotation.minorAllele
- annotation.minorAlleleFreq
- annotation.conservation
- annotation.geneExpression
- annotation.geneTraitAssociation
- annotation.geneDrugInteraction
- annotation.variantTraitAssociation
- annotation.functionalScore
- annotation.additionalAttributes